Structural Basis for the Interaction between the IUS-SPRY Domain of RanBPM and DDX-4 in Germ Cell Development.
Hong, S.K., Kim, K.H., Song, E.J., Kim, E.E.(2016) J Mol Biology 428: 4330-4344
- PubMed: 27622290
- DOI: https://doi.org/10.1016/j.jmb.2016.09.004
- Primary Citation of Related Structures:
5JI7, 5JI9, 5JIA, 5JIU - PubMed Abstract:
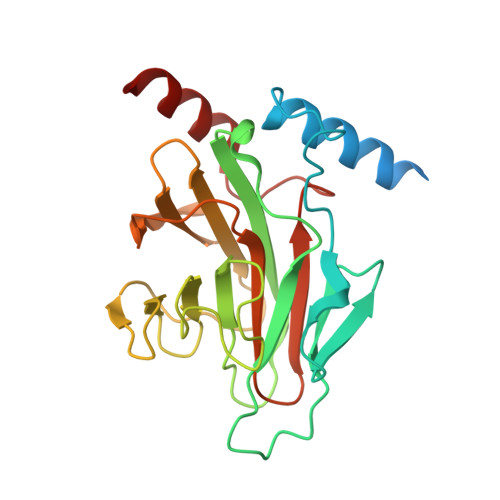
RanBPM and RanBP10 are non-canonical members of the Ran binding protein family that lack the Ran binding domain and do not associate with Ran GTPase in vivo. Rather, they have been shown to be scaffolding proteins that are important for a variety of cellular processes, and both of these proteins contain a SPRY domain, which has been implicated in mediating protein-protein interactions with a variety of targets including the DEAD-box containing ATP-dependent RNA helicase (DDX-4). In this study, we have determined the crystal structures of the SPIa and the ryanodine receptor domain and of approximately 70 upstream residues (immediate upstream to SPRY motif) of both RanBPM and RanBP10. They are almost identical, composed of a β-sandwich fold with a set of two helices on each side located at the edge of the sheets. A unique shallow binding surface is formed by highly conserved loops on the surface of the β-sheet with two aspartates on one end, a positive patch on the opposite end, and a tryptophan lining at the bottom of the surface. The 20-mer peptide (residues 228-247) of human DDX-4, an ATP-dependent RNA helicase known to regulate germ cell development, binds to this surface with a K D of ~13μM. The crystal structure of the peptide complex and the mutagenesis studies elucidate how RanBPM can recognize its interaction partners to function in gametogenesis.
- Biomedical Research Institute, Korea Institute of Science and Technology, Seongbuk-gu Hwarang-ro 14-gil 5, Seoul 02792, Republic of Korea.
Organizational Affiliation: