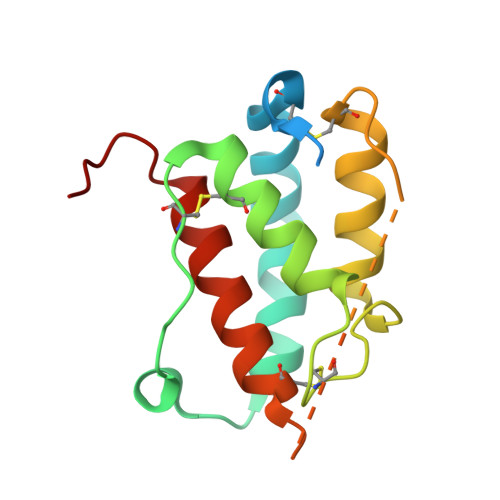
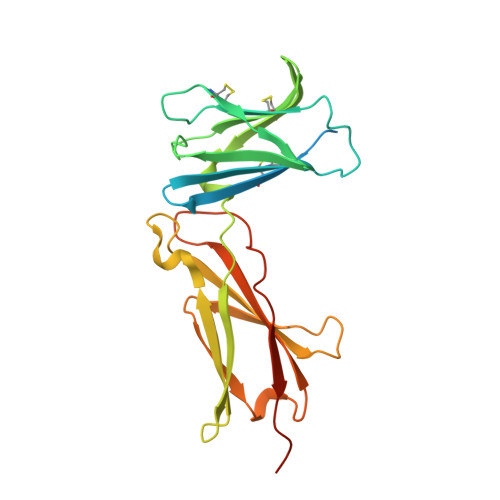
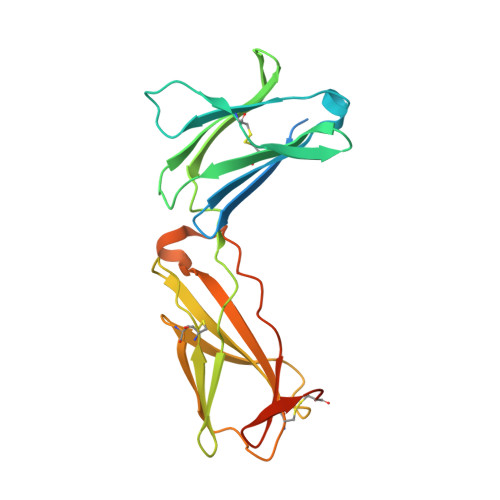
Structure and antagonism of the receptor complex mediated by human TSLP in allergy and asthma.
Verstraete, K., Peelman, F., Braun, H., Lopez, J., Van Rompaey, D., Dansercoer, A., Vandenberghe, I., Pauwels, K., Tavernier, J., Lambrecht, B.N., Hammad, H., De Winter, H., Beyaert, R., Lippens, G., Savvides, S.N.(2017) Nat Commun 8: 14937-14937
- PubMed: 28368013
- DOI: https://doi.org/10.1038/ncomms14937
- Primary Citation of Related Structures:
5J11, 5J13 - PubMed Abstract:
The pro-inflammatory cytokine thymic stromal lymphopoietin (TSLP) is pivotal to the pathophysiology of widespread allergic diseases mediated by type 2 helper T cell (Th2) responses, including asthma and atopic dermatitis. The emergence of human TSLP as a clinical target against asthma calls for maximally harnessing its therapeutic potential via structural and mechanistic considerations. Here we employ an integrative experimental approach focusing on productive and antagonized TSLP complexes and free cytokine. We reveal how cognate receptor TSLPR allosterically activates TSLP to potentiate the recruitment of the shared interleukin 7 receptor α-chain (IL-7Rα) by leveraging the flexibility, conformational heterogeneity and electrostatics of the cytokine. We further show that the monoclonal antibody Tezepelumab partly exploits these principles to neutralize TSLP activity. Finally, we introduce a fusion protein comprising a tandem of the TSLPR and IL-7Rα extracellular domains, which harnesses the mechanistic intricacies of the TSLP-driven receptor complex to manifest high antagonistic potency.
- VIB-UGent Center for Inflammation Research, Zwijnaarde, Ghent 9052, Belgium.
Organizational Affiliation: