Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Ivan, T., Enkvist, E., Viira, B., Manoharan, G.B., Raidaru, G., Pflug, A., Alam, K.A., Zaccolo, M., Engh, R.A., Uri, A.(2016) Bioconjug Chem 27: 1900-1910
- PubMed: 27389935
- DOI: https://doi.org/10.1021/acs.bioconjchem.6b00293
- Primary Citation of Related Structures:
5IZF, 5IZJ, 5J5X - PubMed Abstract:
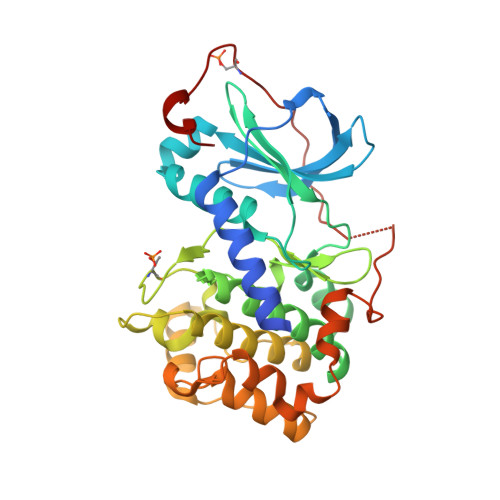
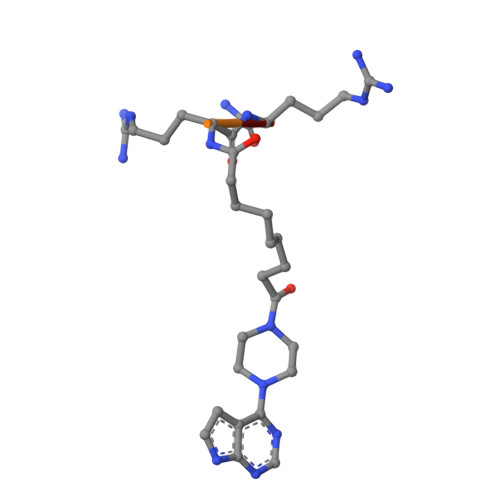
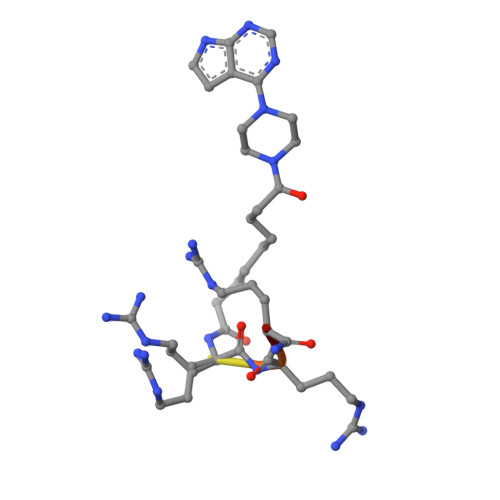
The acknowledged potential of small-molecule therapeutics targeting disease-related protein-protein interactions (PPIs) has promoted active research in this field. The strategy of using small molecule inhibitors (SMIs) to fight strong (tight-binding) PPIs tends to fall short due to the flat and wide interfaces of PPIs. Here we propose a biligand approach for disruption of strong PPIs. The potential of this approach was realized for disruption of the tight-binding (KD = 100 pM) tetrameric holoenzyme of cAMP-dependent protein kinase (PKA). Supported by X-ray analysis of cocrystals, bifunctional inhibitors (ARC-inhibitors) were constructed that simultaneously associated with both the ATP-pocket and the PPI interface area of the catalytic subunit of PKA (PKAc). Bifunctional inhibitor ARC-1411, possessing a KD value of 3 pM toward PKAc, induced the dissociation of the PKA holoenzyme with a low-nanomolar IC50, whereas the ATP-competitive inhibitor H89 bound to the PKA holoenzyme without disruption of the protein tetramer.
- Institute of Chemistry, University of Tartu , 50410 Tartu, Estonia.
Organizational Affiliation: