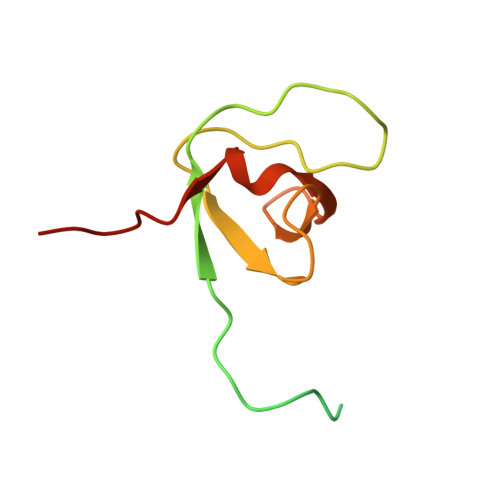
NMR Reveals the Interplay among the AMSH SH3 Binding Motif, STAM2, and Lys63-Linked Diubiquitin.
Hologne, M., Cantrelle, F.X., Riviere, G., Guilliere, F., Trivelli, X., Walker, O.(2016) J Mol Biology 428: 4544-4558
- PubMed: 27725184
- DOI: https://doi.org/10.1016/j.jmb.2016.10.002
- Primary Citation of Related Structures:
5IXF - PubMed Abstract:
AMSH [associated molecule with a Src homology 3 domain of signal transducing adaptor molecule (STAM)] is one of the deubiquitinating enzymes associated in the regulation of endocytic cargo trafficking. It shows an exquisite selectivity for Lys63-linked polyubiquitin chains that are the main chains involved in cargo sorting. The first step requires the ESCRT-0 complex that comprises the STAM and hepatocyte growth factor-regulated substrate (Hrs) proteins. Previous studies have shown that the presence of the STAM protein increases the efficiency of Lys63-linked polyubiquitin chain cleavage by AMSH, one of the deubiquitinating enzyme involved in lysosomal degradation. In the present study, we are seeking to understand if a particular structural organization among these three key players is responsible for the stimulation of the catalytic activity of AMSH. To address this question, we first monitored the interaction between the ubiquitin interacting motif (UIM)-SH3 construct of STAM2 and the Lys63-linked diubiquitin (Lys63-Ub 2 ) chains by means of NMR. We show that Lys63-Ub 2 is able to bind either the UIM or the SH3 domain without any selectivity. We further demonstrate that the SH3 binding motif (SBM) of AMSH (AMSH-SBM) outcompetes Lys63-Ub 2 for binding SH3. Additionally, we show how different AMSH-SBM variants, modified by their sequence and length, exhibit similar equilibrium dissociation constants when binding SH3 but significantly differ in their dissociation rate constants. Finally, we report the solution NMR structure of the AMSH-SBM/SH3 complex and propose a structural organization where the AMSH-SBM interacts with the STAM2-SH3 domain and contributes to the correct positioning of AMSH prior to polyubiquitin chains' cleavage.
- Université de Lyon, CNRS, Université Claude Bernard Lyon1, Ens de Lyon, Institut des Sciences Analytiques, UMR 5280, 5 rue de la Doua, F-69100 Villeurbanne, France.
Organizational Affiliation: