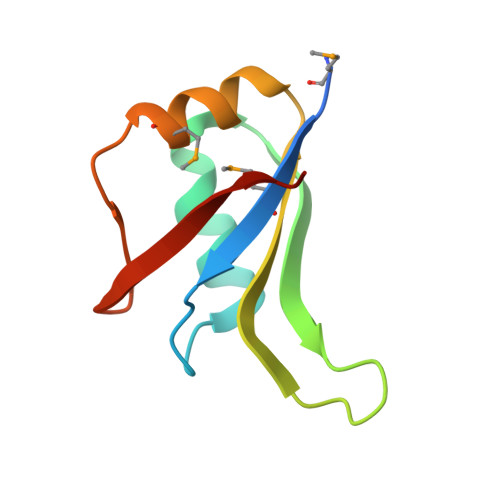
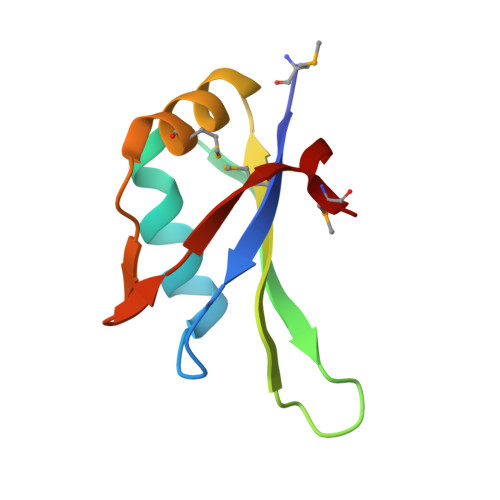
Crystal Structure of the N-Terminal RNA Recognition Motif of mRNA Decay Regulator AUF1.
Choi, Y.J., Yoon, J.H., Chang, J.H.(2016) Biomed Res Int 2016: 3286191-3286191
- PubMed: 27437398
- DOI: https://doi.org/10.1155/2016/3286191
- Primary Citation of Related Structures:
5IM0 - PubMed Abstract:
AU-rich element binding/degradation factor 1 (AUF1) plays a role in destabilizing mRNAs by forming complexes with AU-rich elements (ARE) in the 3'-untranslated regions. Multiple AUF1-ARE complexes regulate the translation of encoded products related to the cell cycle, apoptosis, and inflammation. AUF1 contains two tandem RNA recognition motifs (RRM) and a Gln- (Q-) rich domain in their C-terminal region. To observe how the two RRMs are involved in recognizing ARE, we obtained the AUF1-p37 protein covering the two RRMs. However, only N-terminal RRM (RRM1) was crystallized and its structure was determined at 1.7 Å resolution. It appears that the RRM1 and RRM2 separated before crystallization. To demonstrate which factors affect the separate RRM1-2, we performed limited proteolysis using trypsin. The results indicated that the intact proteins were cleaved by unknown proteases that were associated with them prior to crystallization. In comparison with each of the monomers, the conformations of the β2-β3 loops were highly variable. Furthermore, a comparison with the RRM1-2 structures of HuR and hnRNP A1 revealed that a dimer of RRM1 could be one of the possible conformations of RRM1-2. Our data may provide a guidance for further structural investigations of AUF1 tandem RRM repeat and its mode of ARE binding.
- Department of Biology Education, Kyungpook National University, Daegu 41566, Republic of Korea.
Organizational Affiliation: