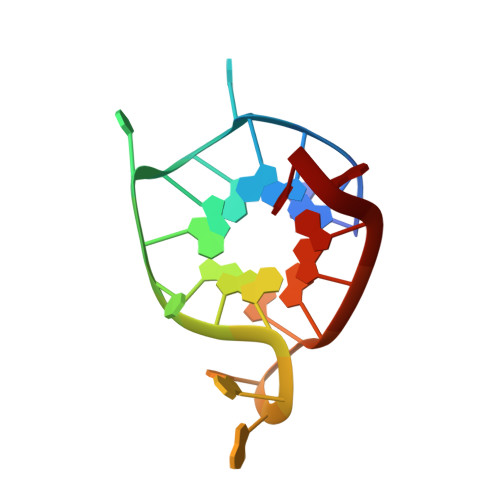
High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation.
Kerkour, A., Marquevielle, J., Ivashchenko, S., Yatsunyk, L.A., Mergny, J.L., Salgado, G.F.(2017) J Biological Chem 292: 8082-8091
- PubMed: 28330874
- DOI: https://doi.org/10.1074/jbc.M117.781906
- Primary Citation of Related Structures:
5I2V - PubMed Abstract:
Non-canonical base pairing within guanine-rich DNA and RNA sequences can produce G-quartets, whose stacking leads to the formation of a G-quadruplex (G4). G4s can coexist with canonical duplex DNA in the human genome and have been suggested to suppress gene transcription, and much attention has therefore focused on studying G4s in promotor regions of disease-related genes. For example, the human KRAS proto-oncogene contains a nuclease-hypersensitive element located upstream of the major transcription start site. The KRAS nuclease-hypersensitive element (NHE) region contains a G-rich element (22RT; 5'-AGGGCGGTGTGGGAATAGGGAA-3') and encompasses a Myc-associated zinc finger-binding site that regulates KRAS transcription. The NEH region therefore has been proposed as a target for new drugs that control KRAS transcription, which requires detailed knowledge of the NHE structure. In this study, we report a high-resolution NMR structure of the G-rich element within the KRAS NHE. We found that the G-rich element forms a parallel structure with three G-quartets connected by a four-nucleotide loop and two short one-nucleotide double-chain reversal loops. In addition, a thymine bulge is found between G8 and G9. The loops of different lengths and the presence of a bulge between the G-quartets are structural elements that potentially can be targeted by small chemical ligands that would further stabilize the structure and interfere or block transcriptional regulators such as Myc-associated zinc finger from accessing their binding sites on the KRAS promoter. In conclusion, our work suggests a possible new route for the development of anticancer agents that could suppress KRAS expression.
- From the Université Bordeaux, INSERM, CNRS, ARNA laboratory, European Institute of Chemistry and Biology, U1212, UMR 5320, 2 Rue Robert Escarpit, 33000 Pessac, France and.
Organizational Affiliation: