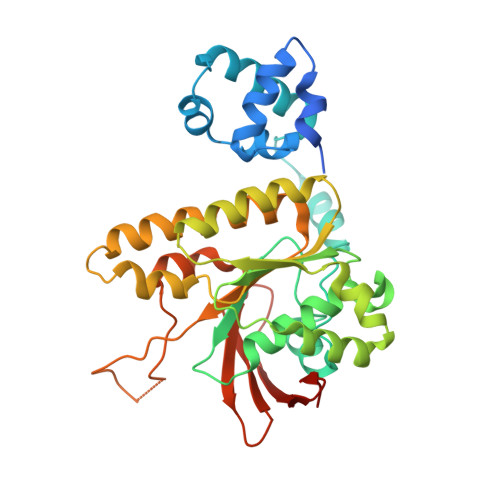
Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Xu, J., Zhao, L., Xu, Y., Zhao, W., Sung, P., Wang, H.W.(2017) Nat Struct Mol Biol 24: 40-46
- PubMed: 27941862
- DOI: https://doi.org/10.1038/nsmb.3336
- Primary Citation of Related Structures:
5H1B, 5H1C - PubMed Abstract:
The central step in eukaryotic homologous recombination (HR) is ATP-dependent DNA-strand exchange mediated by the Rad51 recombinase. In this process, Rad51 assembles on single-stranded DNA (ssDNA) and generates a helical filament that is able to search for and invade homologous double-stranded DNA (dsDNA), thus leading to strand separation and formation of new base pairs between the initiating ssDNA and the complementary strand within the duplex. Here, we used cryo-EM to solve the structures of human RAD51 in complex with DNA molecules, in presynaptic and postsynaptic states, at near-atomic resolution. Our structures reveal both conserved and distinct structural features of the human RAD51-DNA complexes compared with their prokaryotic counterpart. Notably, we also captured the structure of an arrested synaptic complex. Our results provide new insight into the molecular mechanisms of the DNA homology search and strand-exchange processes.
- Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, China.
Organizational Affiliation: