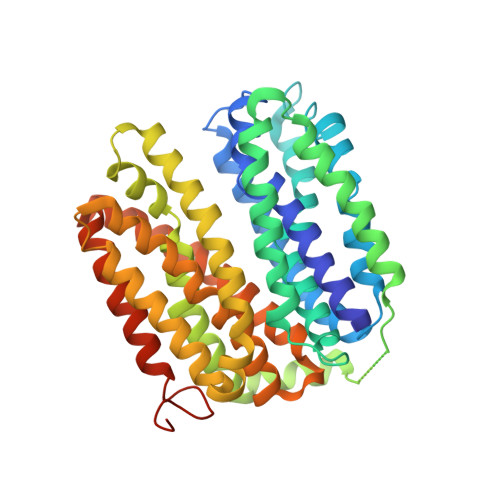
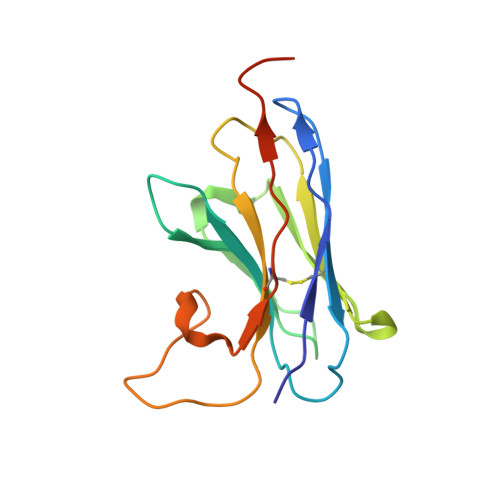
Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation.
Jiang, X., Smirnova, I., Kasho, V., Wu, J., Hirata, K., Ke, M., Pardon, E., Steyaert, J., Yan, N., Kaback, H.R.(2016) Proc Natl Acad Sci U S A 113: 12420-12425
- PubMed: 27791182
- DOI: https://doi.org/10.1073/pnas.1615414113
- Primary Citation of Related Structures:
5GXB - PubMed Abstract:
The lactose permease of Escherichia coli (LacY), a dynamic polytopic membrane protein, catalyzes galactoside-H + symport and operates by an alternating access mechanism that exhibits multiple conformations, the distribution of which is altered by sugar binding. We have developed single-domain camelid nanobodies (Nbs) against a mutant in an outward (periplasmic)-open conformation to stabilize this state of the protein. Here we describe an X-ray crystal structure of a complex between a double-Trp mutant (Gly46→Trp/Gly262→Trp) and an Nb in which free access to the sugar-binding site from the periplasmic cavity is observed. The structure confirms biochemical data indicating that the Nb binds stoichiometrically with nanomolar affinity to the periplasmic face of LacY primarily to the C-terminal six-helix bundle. The structure is novel because the pathway to the sugar-binding site is constricted and the central cavity containing the galactoside-binding site is empty. Although Phe27 narrows the periplasmic cavity, sugar is freely accessible to the binding site. Remarkably, the side chains directly involved in binding galactosides remain in the same position in the absence or presence of bound sugar.
- State Key Laboratory of Membrane Biology, Center for Structural Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: