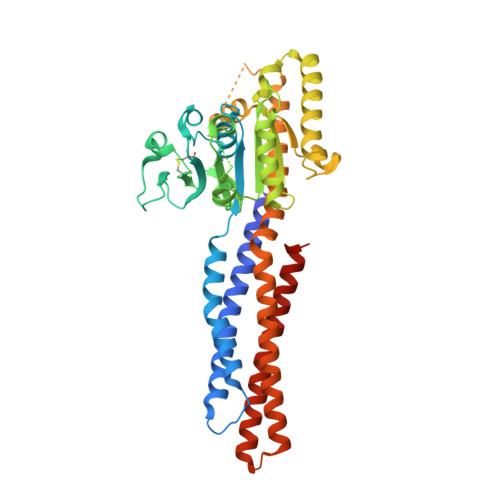
MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion
Cao, Y.L., Meng, S., Chen, Y., Feng, J.X., Gu, D.D., Yu, B., Li, Y.J., Yang, J.Y., Liao, S., Chan, D.C., Gao, S.(2017) Nature 542: 372-376
- PubMed: 28114303
- DOI: https://doi.org/10.1038/nature21077
- Primary Citation of Related Structures:
5GO4, 5GOE, 5GOF, 5GOM - PubMed Abstract:
Mitochondria are double-membraned organelles with variable shapes influenced by metabolic conditions, developmental stage, and environmental stimuli. Their dynamic morphology is a result of regulated and balanced fusion and fission processes. Fusion is crucial for the health and physiological functions of mitochondria, including complementation of damaged mitochondrial DNAs and the maintenance of membrane potential. Mitofusins are dynamin-related GTPases that are essential for mitochondrial fusion. They are embedded in the mitochondrial outer membrane and thought to fuse adjacent mitochondria via combined oligomerization and GTP hydrolysis. However, the molecular mechanisms of this process remain unknown. Here we present crystal structures of engineered human MFN1 containing the GTPase domain and a helical domain during different stages of GTP hydrolysis. The helical domain is composed of elements from widely dispersed sequence regions of MFN1 and resembles the 'neck' of the bacterial dynamin-like protein. The structures reveal unique features of its catalytic machinery and explain how GTP binding induces conformational changes to promote GTPase domain dimerization in the transition state. Disruption of GTPase domain dimerization abolishes the fusogenic activity of MFN1. Moreover, a conserved aspartate residue trigger was found to affect mitochondrial elongation in MFN1, probably through a GTP-loading-dependent domain rearrangement. Thus, we propose a mechanistic model for MFN1-mediated mitochondrial tethering, and our results shed light on the molecular basis of mitochondrial fusion and mitofusin-related human neuromuscular disorders.
- State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou 510060, China.
Organizational Affiliation: