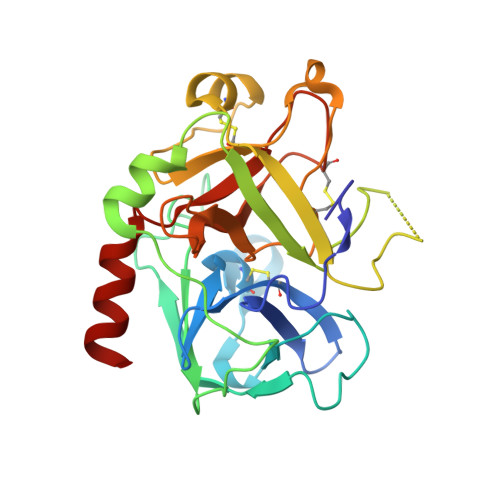
Avathrin: a novel thrombin inhibitor derived from a multicopy precursor in the salivary glands of the ixodid tick, Amblyomma variegatum.
Iyer, J.K., Koh, C.Y., Kazimirova, M., Roller, L., Jobichen, C., Swaminathan, K., Mizuguchi, J., Iwanaga, S., Nuttall, P.A., Chan, M.Y., Kini, R.M.(2017) FASEB J 31: 2981-2995
- PubMed: 28363953
- DOI: https://doi.org/10.1096/fj.201601216R
- Primary Citation of Related Structures:
5GIM - PubMed Abstract:
Tick saliva is a rich source of antihemostatic compounds. We amplified a cDNA from the salivary glands of the tropical bont tick ( Amblyomma variegatum ) using primers based on the variegin sequence, which we previously identified as a novel thrombin inhibitor from the same tick species. The transcript encodes a precursor protein comprising a signal peptide and 5 repeats of variegin-like sequences that could be processed into multiple short peptides. These peptides share 31 to 34% identity with variegin. Here, we structurally and functionally characterized one of these peptides named "avathrin." Avathrin is a fast, tight binding competitive inhibitor with an affinity of 545 pM for thrombin and is 4 orders of magnitude more selective towards thrombin than to the other serine proteases of the coagulation cascade. The crystal structure of thrombin-avathrin complex at 2.09 Å revealed that avathrin interacts with the thrombin active site and exosite-I. Although avathrin is cleaved by thrombin, the C-terminal cleavage product continues to exert prolonged inhibition. Avathrin is more potent than hirulog-1 in a murine carotid artery thrombosis model. Such precursor proteins that could be processed into multiple thrombin inhibiting peptides appear to be widespread among Amblyomminae, providing an enormous library of molecules for development as potent antithrombotics.-Iyer, J. K., Koh, C. Y., Kazimirova, M., Roller, L., Jobichen, C., Swaminathan, K., Mizuguchi, J., Iwanaga, S., Nuttall, P. A., Chan, M. Y., Kini, R. M. Avathrin: a novel thrombin inhibitor derived from a multicopy precursor in the salivary glands of the ixodid tick, Amblyomma variegatum .
- Protein Science Laboratory, Department of Biological Sciences, National University of Singapore, Singapore.
Organizational Affiliation: