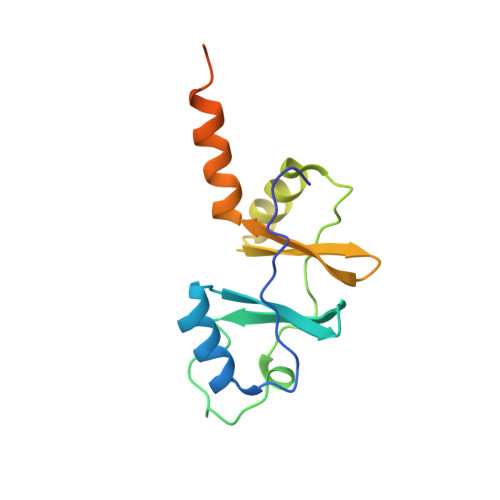
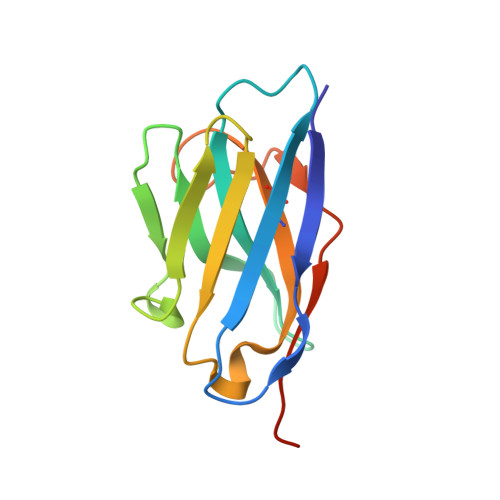
Crystallographic and biochemical characterization of the dimeric architecture of site-2 protease.
Schacherl, M., Gompert, M., Pardon, E., Lamkemeyer, T., Steyaert, J., Baumann, U.(2017) Biochim Biophys Acta 1859: 1859-1871
- PubMed: 28502790
- DOI: https://doi.org/10.1016/j.bbamem.2017.05.006
- Primary Citation of Related Structures:
5G5R, 5G5X - PubMed Abstract:
Regulated intramembrane proteolysis by members of the site-2 protease family (S2P) is an essential signal transduction mechanism conserved from bacteria to humans. There is some evidence that extra-membranous domains, like PDZ and CBS domains, regulate the proteolytic activity of S2Ps and that some members act as dimers. Here we report the crystal structure of the regulatory CBS domain pair of S2P from Archaeoglobus fulgidus, AfS2P, in the apo and nucleotide-bound form in complex with a specific nanobody from llama. Cross-linking and SEC-MALS analyses show for the first time the dimeric architecture of AfS2P both in the membrane and in detergent micelles. The CBS domain pair dimer (CBS module) displays an unusual head-to-tail configuration and nucleotide binding triggers no major conformational changes in the magnesium-free state. In solution, MgATP drives monomerization of the CBS module. We propose a model of the so far unknown architecture of the transmembrane domain dimer and for a regulatory mechanism of AfS2P that involves the interaction of positively charged arginine residues located at the cytoplasmic face of the transmembrane domain with the negatively charged phosphate groups of ATP moieties bound to the CBS domain pairs. Binding of MgATP could promote opening of the CBS module to allow lateral access of the globular cytoplasmic part of the substrate.
- Institute of Biochemistry, University of Cologne, Otto-Fischer-Str. 14, 50674 Cologne, Germany. Electronic address: magdalena.schacherl@uni-koeln.de.
Organizational Affiliation: