Mode of Action Uncovered for the Specific Reduction of Methane Emissions from Ruminants by the Small Molecule 3-Nitrooxypropanol.
Duin, E.C., Wagner, T., Shima, S., Prakash, D., Cronin, B., Yanez-Ruiz, D.R., Duval, S., Rumbeli, R., Stemmler, R.T., Thauer, R.K., Kindermann, M.(2016) Proc Natl Acad Sci U S A 113: 6172
- PubMed: 27140643
- DOI: https://doi.org/10.1073/pnas.1600298113
- Primary Citation of Related Structures:
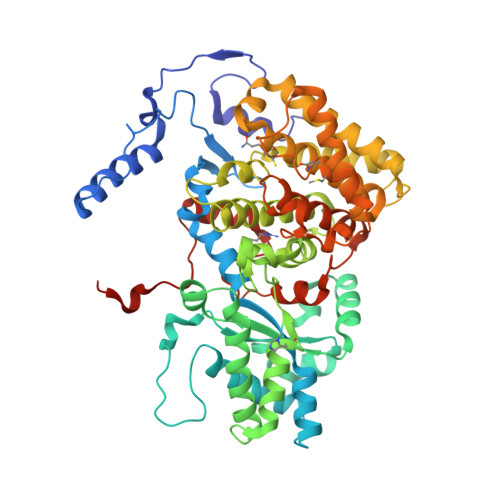
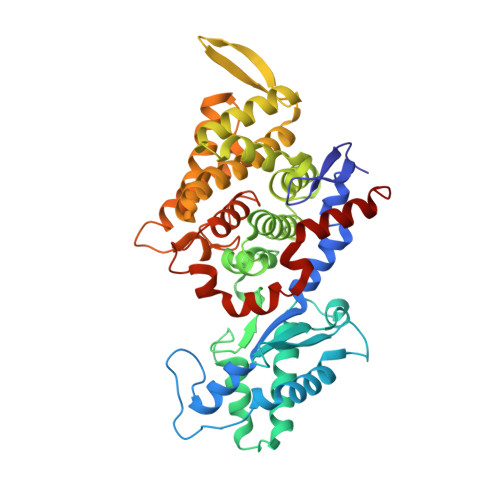
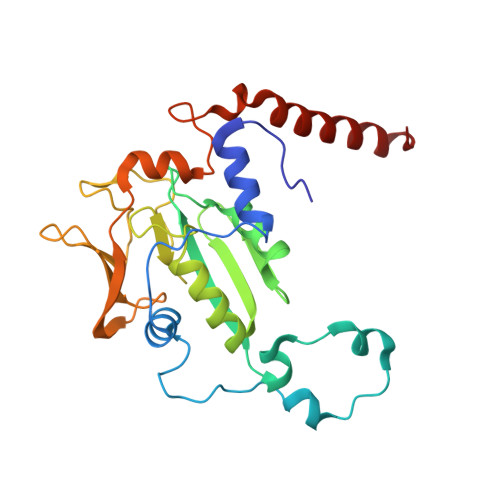
5G0R - PubMed Abstract:
Ruminants, such as cows, sheep, and goats, predominantly ferment in their rumen plant material to acetate, propionate, butyrate, CO2, and methane. Whereas the short fatty acids are absorbed and metabolized by the animals, the greenhouse gas methane escapes via eructation and breathing of the animals into the atmosphere. Along with the methane, up to 12% of the gross energy content of the feedstock is lost. Therefore, our recent report has raised interest in 3-nitrooxypropanol (3-NOP), which when added to the feed of ruminants in milligram amounts persistently reduces enteric methane emissions from livestock without apparent negative side effects [Hristov AN, et al. (2015) Proc Natl Acad Sci USA 112(34):10663-10668]. We now show with the aid of in silico, in vitro, and in vivo experiments that 3-NOP specifically targets methyl-coenzyme M reductase (MCR). The nickel enzyme, which is only active when its Ni ion is in the +1 oxidation state, catalyzes the methane-forming step in the rumen fermentation. Molecular docking suggested that 3-NOP preferably binds into the active site of MCR in a pose that places its reducible nitrate group in electron transfer distance to Ni(I). With purified MCR, we found that 3-NOP indeed inactivates MCR at micromolar concentrations by oxidation of its active site Ni(I). Concomitantly, the nitrate ester is reduced to nitrite, which also inactivates MCR at micromolar concentrations by oxidation of Ni(I). Using pure cultures, 3-NOP is demonstrated to inhibit growth of methanogenic archaea at concentrations that do not affect the growth of nonmethanogenic bacteria in the rumen.
- Department of Chemistry and Biochemistry, Auburn University, Auburn, AL 36849;
Organizational Affiliation: