Structure of the Dispase Autolysis Inducing Protein from Streptomyces Mobaraensis and Glutamine Cross-Linking Sites for Transglutaminase
Fiebig, D., Schmelz, S., Zindel, S., Ehret, V., Beck, J., Ebenig, A., Ehret, M., Froels, S., Pfeifer, F., Kolmar, H., Fuchsbauer, H.L., Scrima, A.(2016) J Biological Chem 291: 20417
- PubMed: 27493205
- DOI: https://doi.org/10.1074/jbc.M116.731109
- Primary Citation of Related Structures:
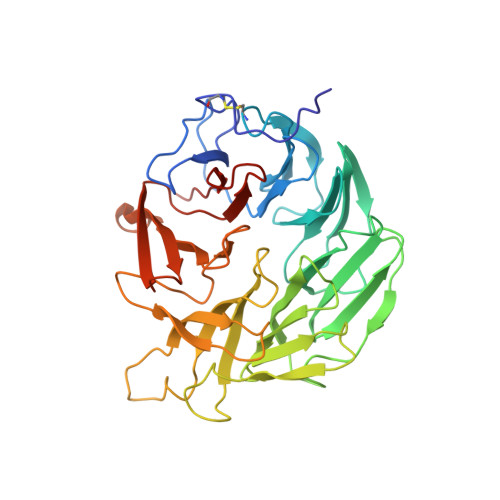
5FZP - PubMed Abstract:
Transglutaminase from Streptomyces mobaraensis (MTG) is an important enzyme for cross-linking and modifying proteins. An intrinsic substrate of MTG is the dispase autolysis-inducing protein (DAIP). The amino acid sequence of DAIP contains 5 potential glutamines and 10 lysines for MTG-mediated cross-linking. The aim of the study was to determine the structure and glutamine cross-linking sites of the first physiological MTG substrate. A production procedure was established in Escherichia coli BL21 (DE3) to obtain high yields of recombinant DAIP. DAIP variants were prepared by replacing four of five glutamines for asparagines in various combinations via site-directed mutagenesis. Incorporation of biotin cadaverine revealed a preference of MTG for the DAIP glutamines in the order of Gln-39 ≫ Gln-298 > Gln-345 ∼ Gln-65 ≫ Gln-144. In the structure of DAIP the preferred glutamines do cluster at the top of the seven-bladed β-propeller. This suggests a targeted cross-linking of DAIP by MTG that may occur after self-assembly in the bacterial cell wall. Based on our biochemical and structural data of the first physiological MTG substrate, we further provide novel insight into determinants of MTG-mediated modification, specificity, and efficiency.
- Department of Chemical Engineering and Biotechnology, University of Applied Sciences of Darmstadt, 64287 Darmstadt, Germany, and the Department of Chemistry and.
Organizational Affiliation: