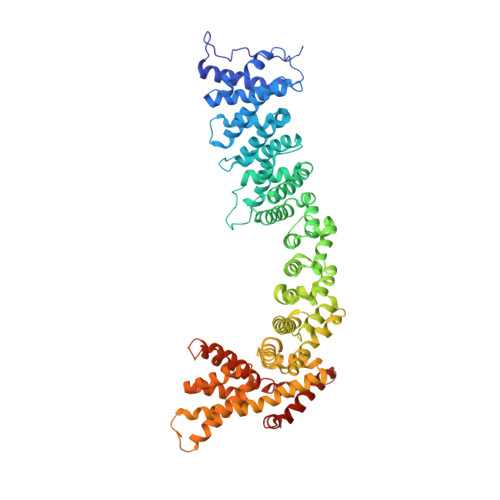
Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Muir, K.W., Kschonsak, M., Li, Y., Metz, J., Haering, C.H., Panne, D.(2016) Cell Rep 14: 2116
- PubMed: 26923589
- DOI: https://doi.org/10.1016/j.celrep.2016.01.078
- Primary Citation of Related Structures:
5FRP, 5FRR, 5FRS - PubMed Abstract:
Sister chromatid cohesion is a fundamental prerequisite to faithful genome segregation. Cohesion is precisely regulated by accessory factors that modulate the stability with which the cohesin complex embraces chromosomes. One of these factors, Pds5, engages cohesin through Scc1 and is both a facilitator of cohesion, and, conversely also mediates the release of cohesin from chromatin. We present here the crystal structure of a complex between budding yeast Pds5 and Scc1, thus elucidating the molecular basis of Pds5 function. Pds5 forms an elongated HEAT repeat that binds to Scc1 via a conserved surface patch. We demonstrate that the integrity of the Pds5-Scc1 interface is indispensable for the recruitment of Pds5 to cohesin, and that its abrogation results in loss of sister chromatid cohesion and cell viability.
- European Molecular Biology Laboratory Grenoble Outstation and Unit of Virus Host-Cell Interactions, University Grenoble Alpes-CNRS-EMBL, 71 Avenue des Martyrs, CS 90181, 38042 Grenoble Cedex 9, France.
Organizational Affiliation: