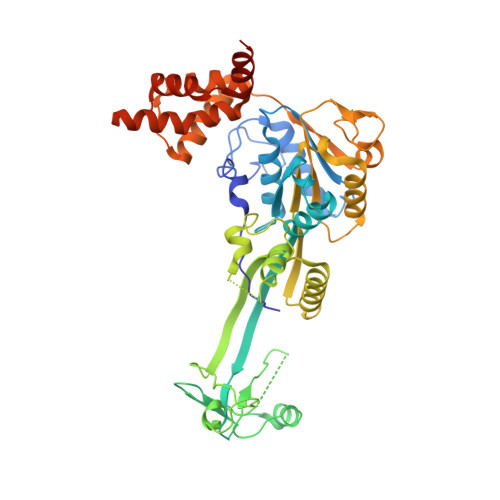
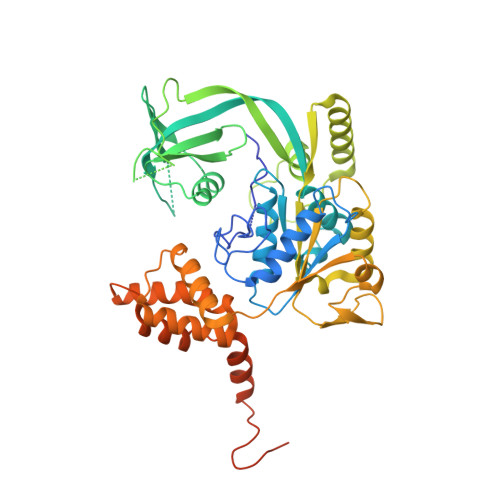
The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Silva-Martin, N., Dauden, M.I., Glatt, S., Hoffmann, N.A., Kastritis, P., Bork, P., Beck, M., Muller, C.W.(2016) PLoS One 11: 46457
- PubMed: 26745716
- DOI: https://doi.org/10.1371/journal.pone.0146457
- Primary Citation of Related Structures:
5FM6, 5FM7 - PubMed Abstract:
The Rvb1/Rvb2 complex is an essential component of many cellular pathways. The Rvb1/Rvb2 complex forms a dodecameric assembly where six copies of each subunit form two heterohexameric rings. However, due to conformational variability, the way the two rings pack together is still not fully understood. Here, we present the crystal structure and two cryo-electron microscopy reconstructions of the dodecameric, full-length Rvb1/Rvb2 complex, all showing that the interaction between the two heterohexameric rings is mediated through the Rvb1/Rvb2-specific domain II. Two conformations of the Rvb1/Rvb2 dodecamer are present in solution: a stretched conformation also present in the crystal, and a compact conformation. Novel asymmetric features observed in the reconstruction of the compact conformation provide additional insight into the plasticity of the Rvb1/Rvb2 complex.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany.
Organizational Affiliation: