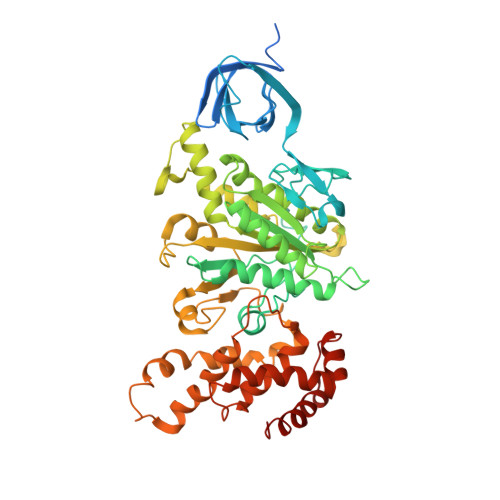
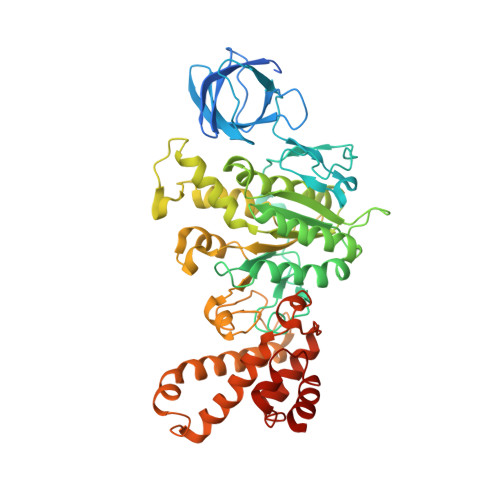
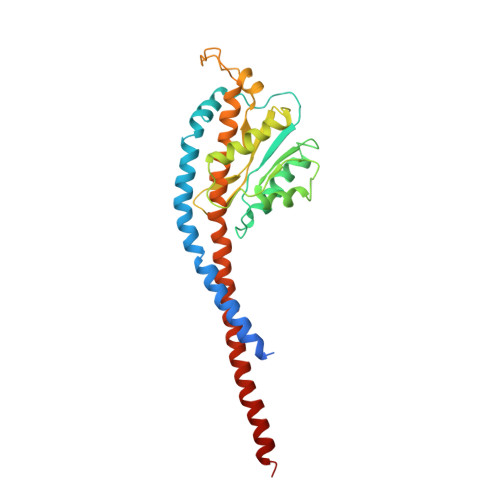
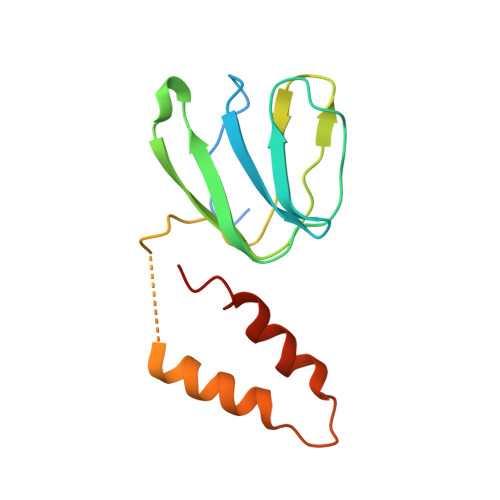
Structure of a Complete ATP Synthase Dimer Reveals the Molecular Basis of Inner Mitochondrial Membrane Morphology.
Hahn, A., Parey, K., Bublitz, M., Mills, D.J., Zickermann, V., Vonck, J., Kuhlbrandt, W., Meier, T.(2016) Mol Cell 63: 445
- PubMed: 27373333
- DOI: https://doi.org/10.1016/j.molcel.2016.05.037
- Primary Citation of Related Structures:
5FL7 - PubMed Abstract:
We determined the structure of a complete, dimeric F1Fo-ATP synthase from yeast Yarrowia lipolytica mitochondria by a combination of cryo-EM and X-ray crystallography. The final structure resolves 58 of the 60 dimer subunits. Horizontal helices of subunit a in Fo wrap around the c-ring rotor, and a total of six vertical helices assigned to subunits a, b, f, i, and 8 span the membrane. Subunit 8 (A6L in human) is an evolutionary derivative of the bacterial b subunit. On the lumenal membrane surface, subunit f establishes direct contact between the two monomers. Comparison with a cryo-EM map of the F1Fo monomer identifies subunits e and g at the lateral dimer interface. They do not form dimer contacts but enable dimer formation by inducing a strong membrane curvature of ∼100°. Our structure explains the structural basis of cristae formation in mitochondria, a landmark signature of eukaryotic cell morphology.
- Department of Structural Biology, Max Planck Institute of Biophysics, Max-von-Laue-Str. 3, 60438 Frankfurt am Main, Germany.
Organizational Affiliation: