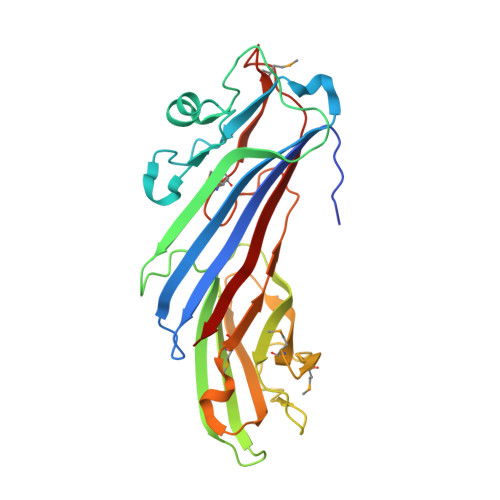
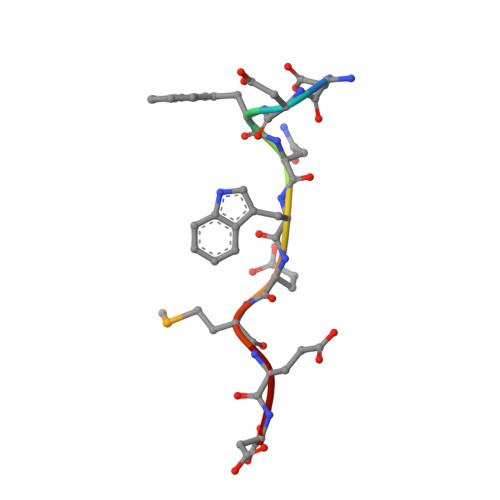
Structural Basis for the Binding of Tryptophan-Based Motifs by Delta-Cop.
Suckling, R.J., Poon, P.P., Travis, S.M., Majoul, I.V., Hughson, F.M., Evans, P.R., Duden, R., Owen, D.J.(2015) Proc Natl Acad Sci U S A 112: 14242
- PubMed: 26578768
- DOI: https://doi.org/10.1073/pnas.1506186112
- Primary Citation of Related Structures:
5FJW, 5FJX, 5FJZ, 5FK0 - PubMed Abstract:
Coatomer consists of two subcomplexes: the membrane-targeting, ADP ribosylation factor 1 (Arf1):GTP-binding βγδζ-COP F-subcomplex, which is related to the adaptor protein (AP) clathrin adaptors, and the cargo-binding αβ'ε-COP B-subcomplex. We present the structure of the C-terminal μ-homology domain of the yeast δ-COP subunit in complex with the WxW motif from its binding partner, the endoplasmic reticulum-localized Dsl1 tether. The motif binds at a site distinct from that used by the homologous AP μ subunits to bind YxxΦ cargo motifs with its two tryptophan residues sitting in compatible pockets. We also show that the Saccharomyces cerevisiae Arf GTPase-activating protein (GAP) homolog Gcs1p uses a related WxxF motif at its extreme C terminus to bind to δ-COP at the same site in the same way. Mutations designed on the basis of the structure in conjunction with isothermal titration calorimetry confirm the mode of binding and show that mammalian δ-COP binds related tryptophan-based motifs such as that from ArfGAP1 in a similar manner. We conclude that δ-COP subunits bind Wxn(1-6)[WF] motifs within unstructured regions of proteins that influence the lifecycle of COPI-coated vesicles; this conclusion is supported by the observation that, in the context of a sensitizing domain deletion in Dsl1p, mutating the tryptophan-based motif-binding site in yeast causes defects in both growth and carboxypeptidase Y trafficking/processing.
- Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom; Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 0XY, United Kingdom;
Organizational Affiliation: