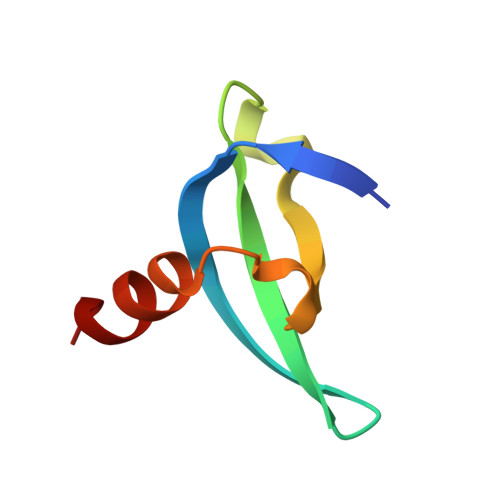
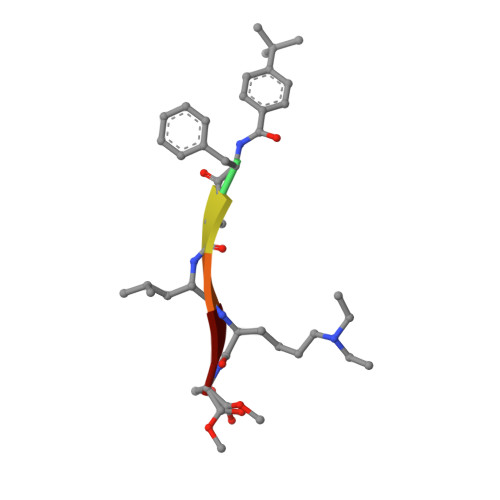
A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Stuckey, J.I., Dickson, B.M., Cheng, N., Liu, Y., Norris, J.L., Cholensky, S.H., Tempel, W., Qin, S., Huber, K.G., Sagum, C., Black, K., Li, F., Huang, X.P., Roth, B.L., Baughman, B.M., Senisterra, G., Pattenden, S.G., Vedadi, M., Brown, P.J., Bedford, M.T., Min, J., Arrowsmith, C.H., James, L.I., Frye, S.V.(2016) Nat Chem Biol 12: 180-187
- PubMed: 26807715
- DOI: https://doi.org/10.1038/nchembio.2007
- Primary Citation of Related Structures:
5EPK, 5EPL, 5EQ0 - PubMed Abstract:
We report the design and characterization of UNC3866, a potent antagonist of the methyllysine (Kme) reading function of the Polycomb CBX and CDY families of chromodomains. Polycomb CBX proteins regulate gene expression by targeting Polycomb repressive complex 1 (PRC1) to sites of H3K27me3 via their chromodomains. UNC3866 binds the chromodomains of CBX4 and CBX7 most potently, with a K(d) of ∼100 nM for each, and is 6- to 18-fold selective as compared to seven other CBX and CDY chromodomains while being highly selective over >250 other protein targets. X-ray crystallography revealed that UNC3866's interactions with the CBX chromodomains closely mimic those of the methylated H3 tail. UNC4195, a biotinylated derivative of UNC3866, was used to demonstrate that UNC3866 engages intact PRC1 and that EED incorporation into PRC1 is isoform dependent in PC3 prostate cancer cells. Finally, UNC3866 inhibits PC3 cell proliferation, consistent with the known ability of CBX7 overexpression to confer a growth advantage, whereas UNC4219, a methylated negative control compound, has negligible effects.
- Center for Integrative Chemical Biology and Drug Discovery, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Organizational Affiliation: