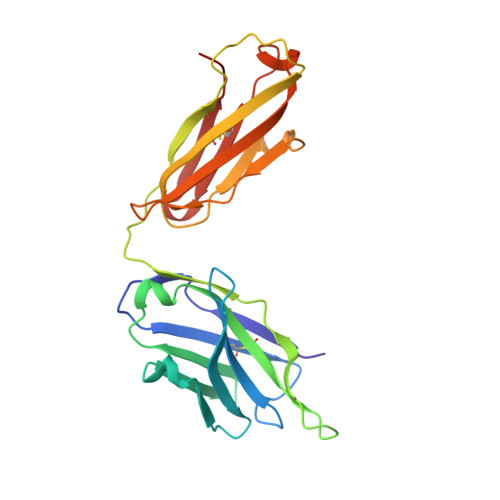
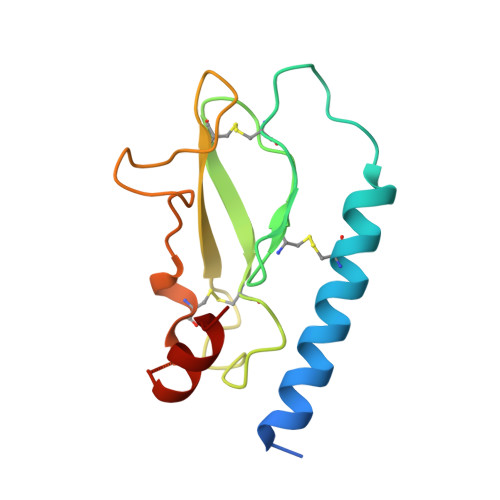
Structural insight into antibody-mediated antagonism of the Glucagon-like peptide-1 Receptor.
Hennen, S., Kodra, J.T., Soroka, V., Krogh, B.O., Wu, X., Kaastrup, P., Orskov, C., Ronn, S.G., Schluckebier, G., Barbateskovic, S., Gandhi, P.S., Reedtz-Runge, S.(2016) Sci Rep 6: 26236-26236
- PubMed: 27196125
- DOI: https://doi.org/10.1038/srep26236
- Primary Citation of Related Structures:
5E94 - PubMed Abstract:
The Glucagon-like peptide-1 receptor (GLP-1R) is a member of the class B G protein-coupled receptor (GPCR) family and a well-established target for the treatment of type 2 diabetes. The N-terminal extracellular domain (ECD) of GLP-1R is important for GLP-1 binding and the crystal structure of the GLP-1/ECD complex was reported previously. The first structure of a class B GPCR transmembrane (TM) domain was solved recently, but the full length receptor structure is still not well understood. Here we describe the molecular details of antibody-mediated antagonism of the GLP-1R using both in vitro pharmacology and x-ray crystallography. We showed that the antibody Fab fragment (Fab 3F52) blocked the GLP-1 binding site of the ECD directly and thereby acts as a competitive antagonist of native GLP-1. Interestingly, Fab 3F52 also blocked a short peptide agonist believed to engage primarily the transmembrane and extracellular loop region of GLP-1R, whereas functionality of an allosteric small-molecule agonist was not inhibited. This study has implications for the structural understanding of the GLP-1R and related class B GPCRs, which is important for the development of new and improved therapeutics targeting these receptors.
- Incretin Biology, Novo Nordisk, Novo Nordisk Park, DK-2760, Måløv, Denmark.
Organizational Affiliation: