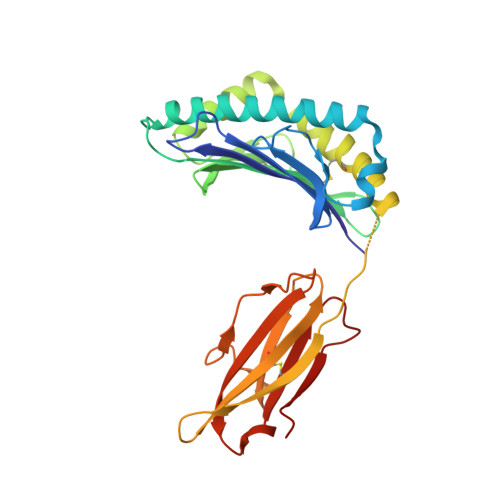
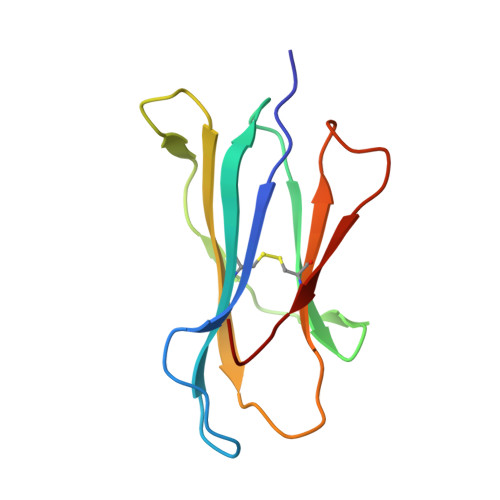
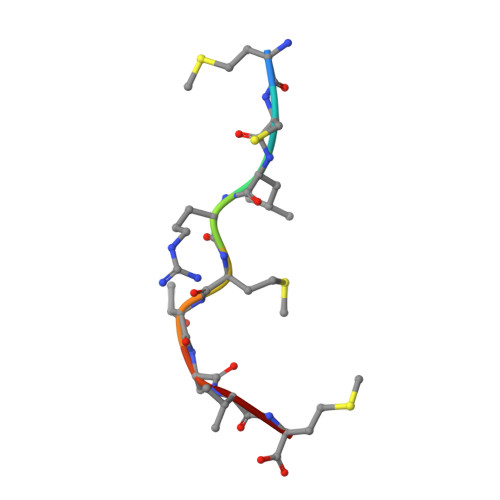
The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
Hafstrand, I., Doorduijn, E.M., Duru, A.D., Buratto, J., Oliveira, C.C., Sandalova, T., van Hall, T., Achour, A.(2016) J Immunol 196: 2327-2334
- PubMed: 26800871
- DOI: https://doi.org/10.4049/jimmunol.1502249
- Primary Citation of Related Structures:
5E8N, 5E8O, 5E8P - PubMed Abstract:
MHC class I downregulation represents a significant challenge for successful T cell-based immunotherapy. T cell epitopes associated with impaired peptide processing (TEIPP) constitute a novel category of immunogenic Ags that are selectively presented on transporter associated with Ag processing-deficient cells. The TEIPP neoepitopes are CD8 T cell targets, derived from nonmutated self-proteins that might be exploited to prevent immune escape. In this study, the crystal structure of H-2D(b) in complex with the first identified TEIPP Ag (MCLRMTAVM) derived from the Trh4 protein has been determined to 2.25 Å resolution. In contrast to prototypic H-2D(b) peptides, Trh4 takes a noncanonical peptide-binding pattern with extensive sulfur-π interactions that contribute to the overall complex stability. Importantly, the noncanonical methionine at peptide position 5 acts as a main anchor, altering only the conformation of the H-2D(b) residues Y156 and H155 and thereby forming a unique MHC/peptide conformer that is essential for recognition by TEIPP-specific T cells. Substitution of peptide residues p2C and p5M to the conservative α-aminobutyric acid and norleucine, respectively, significantly reduced complex stability, without altering peptide conformation or T cell recognition. In contrast, substitution of p5M to a conventional asparagine abolished recognition by the H-2D(b)/Trh4-specific T cell clone LnB5. We anticipate that the H-2D(b)/Trh4 complex represents the first example, to our knowledge, of a broader repertoire of alternative MHC class I binders.
- Science for Life Laboratory, Department of Medicine, Solna, Karolinska Institutet, SE-10450 Stockholm, Sweden; and.
Organizational Affiliation: