Crystal structure of TCR PF8 in complex with flu MP(58-66) epitope presented by HLA-A2
Gao, M.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TCR alpha chain,Human nkt tcr alpha chain | A [auth G], F [auth A], K [auth F], P | 208 | Homo sapiens | Mutation(s): 0 | 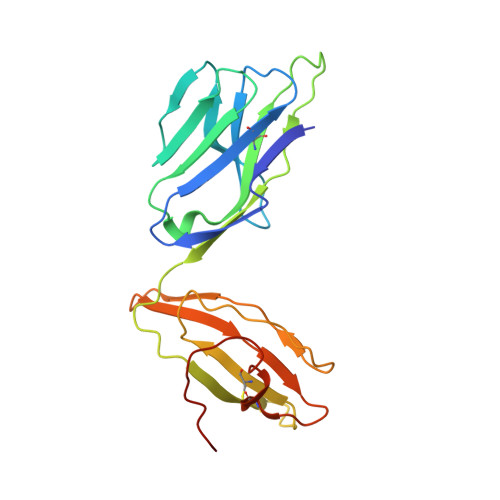 |
UniProt | |||||
Find proteins for K7N5M3 (Homo sapiens) Explore K7N5M3 Go to UniProtKB: K7N5M3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | K7N5M3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T-cell receptor beta-2 chain C region | B [auth H], G [auth B], L, Q | 243 | Homo sapiens | Mutation(s): 0 Gene Names: TRBC2 | 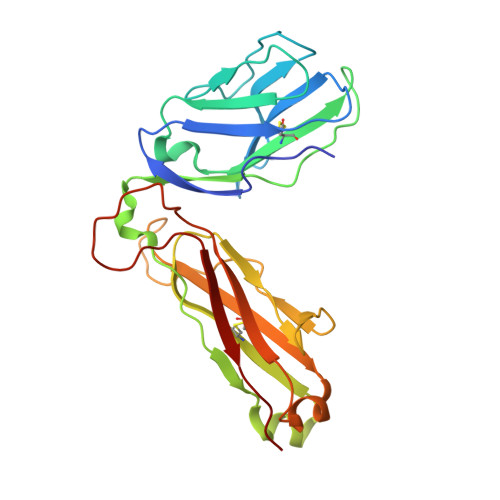 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HLA class I histocompatibility antigen, A-2 alpha chain | C [auth I], H [auth C], M, R | 276 | Homo sapiens | Mutation(s): 0 Gene Names: HLA-A, HLAA | 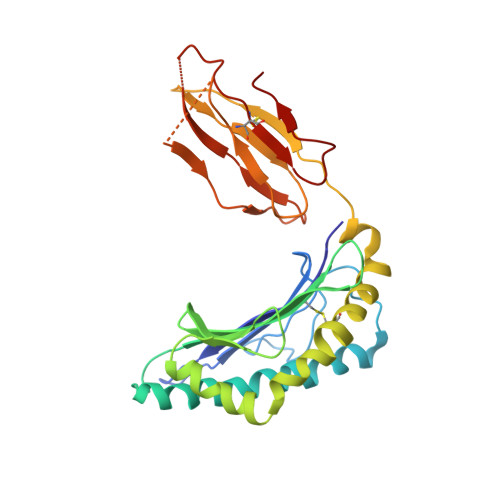 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04439 (Homo sapiens) Explore P04439 Go to UniProtKB: P04439 | |||||
PHAROS: P04439 GTEx: ENSG00000206503 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04439 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | D [auth J], I [auth D], N, S [auth U] | 100 | Homo sapiens | Mutation(s): 0 Gene Names: B2M, CDABP0092, HDCMA22P | 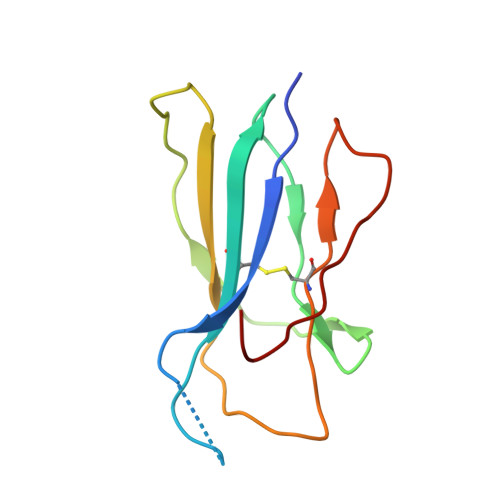 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Matrix protein 1 | E [auth K], J [auth E], O, T | 9 | Influenza A virus (A/Boston/2747243M/2009(H1N1)) | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P03485 (Influenza A virus (strain A/Puerto Rico/8/1934 H1N1)) Explore P03485 Go to UniProtKB: P03485 | |||||
Entity Groups | |||||
| UniProt Group | P03485 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 71.853 | α = 83.66 |
| b = 123.614 | β = 74.87 |
| c = 139.036 | γ = 73.17 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| d*TREK | data scaling |
| PHASER | phasing |
| Coot | model building |
| PDB_EXTRACT | data extraction |
| CrystalClear | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | AI036900 |