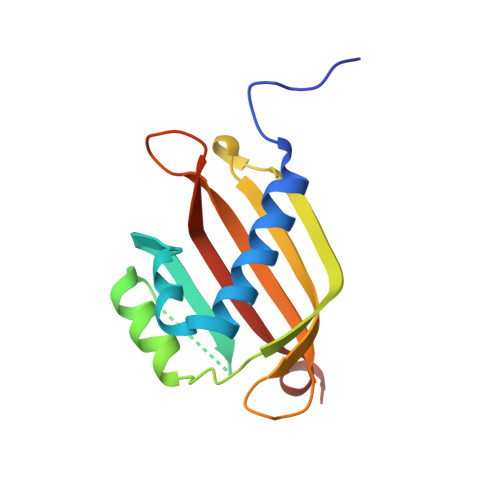
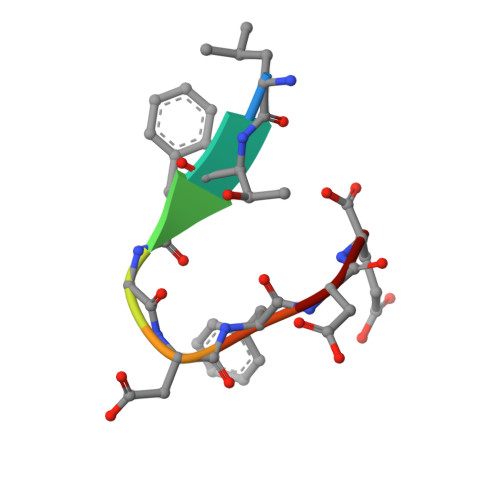
Crystal structure of the G3BP2 NTF2-like domain in complex with a canonical FGDF motif peptide.
Kristensen, O.(2015) Biochem Biophys Res Commun 467: 53-57
- PubMed: 26410532
- DOI: https://doi.org/10.1016/j.bbrc.2015.09.123
- Primary Citation of Related Structures:
5DRV - PubMed Abstract:
The crystal structure of the NTF2-like domain of the human Ras GTPase SH3 Binding Protein (G3BP), isoform 2, was determined at a resolution of 2.75 Å in complex with a peptide containing a FGDF sequence motif. The overall structure of the protein is highly similar to the homodimeric N-terminal domains of the G3BP1 and Rasputin proteins. Recently, a subset of G3BP interacting proteins was recognized to share a common sequence motif, FGDF. The most studied binding partners, USP10 and viral nsP3, interfere with essential G3BP functions related to assembly of cellular stress granules. Reported molecular modeling suggested that FGDF-motif containing peptides bind in an extended conformation into a hydrophobic groove on the surface of the G3BP NTF2-like domain in a manner similar to the known binding of FxFG nucleoporin repeats. The results in this paper provide evidence for a different binding mode. The FGDF peptide binds and changes conformation of the protruding N-terminal residues by providing hydrophobic interactions to a symmetry related molecule that facilitated crystallization of the G3BP2 isoform.
- Biostructural Research, Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Universitetsparken 2, 2100 Copenhagen, Denmark. Electronic address: ok@sund.ku.dk.
Organizational Affiliation: