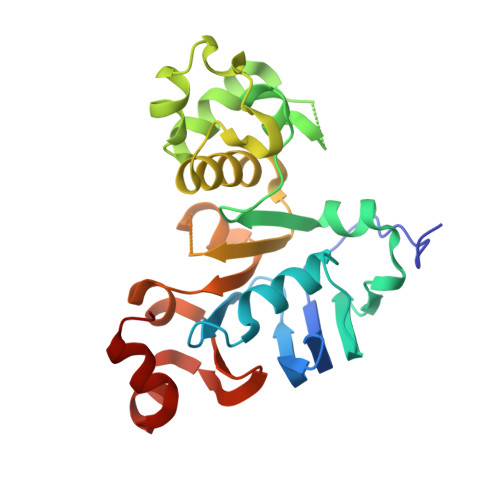
1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
Halavaty, A.S., Minasov, G., Dubrovska, I., Kiryukhina, O., Grimshaw, S., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.