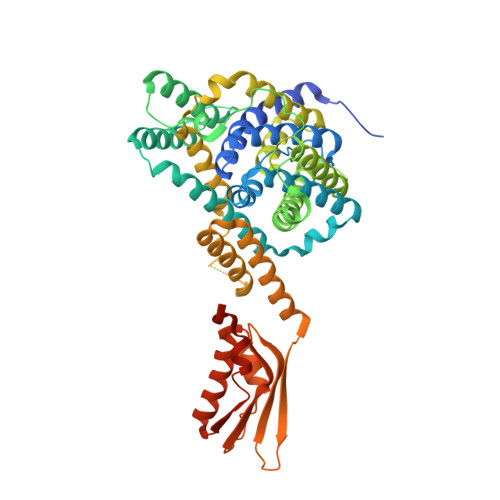
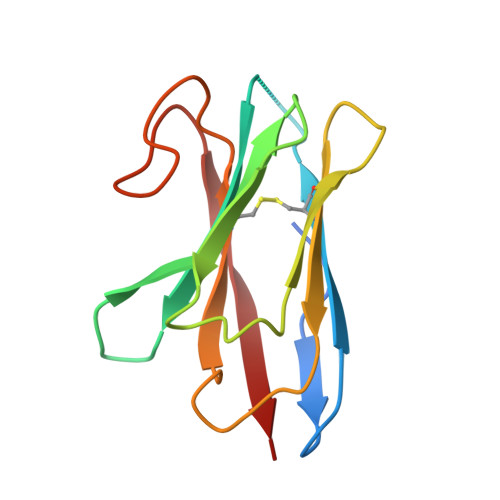
Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family.
Geertsma, E.R., Chang, Y.N., Shaik, F.R., Neldner, Y., Pardon, E., Steyaert, J., Dutzler, R.(2015) Nat Struct Mol Biol 22: 803-808
- PubMed: 26367249
- DOI: https://doi.org/10.1038/nsmb.3091
- Primary Citation of Related Structures:
5DA0, 5DA4, 5IOF - PubMed Abstract:
The SLC26 family of membrane proteins combines a variety of functions within a conserved molecular scaffold. Its members, besides coupled anion transporters and channels, include the motor protein Prestin, which confers electromotility to cochlear outer hair cells. To gain insight into the architecture of this protein family, we characterized the structure and function of SLC26Dg, a facilitator of proton-coupled fumarate symport, from the bacterium Deinococcus geothermalis. Its modular structure combines a transmembrane unit and a cytoplasmic STAS domain. The membrane-inserted domain consists of two intertwined inverted repeats of seven transmembrane segments each and resembles the fold of the unrelated transporter UraA. It shows an inward-facing, ligand-free conformation with a potential substrate-binding site at the interface between two helix termini at the center of the membrane. This structure defines the common framework for the diverse functional behavior of the SLC26 family.
- Department of Biochemistry, University of Zurich, Zurich, Switzerland.
Organizational Affiliation: