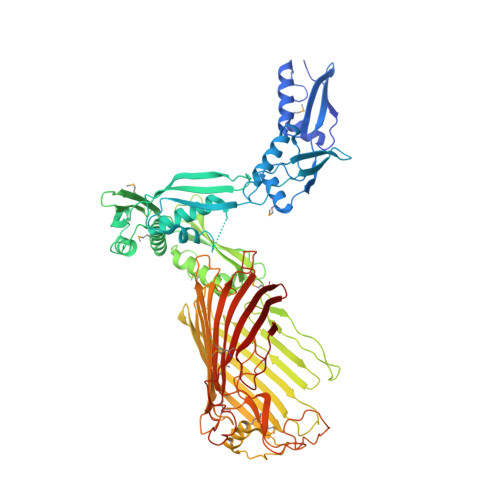
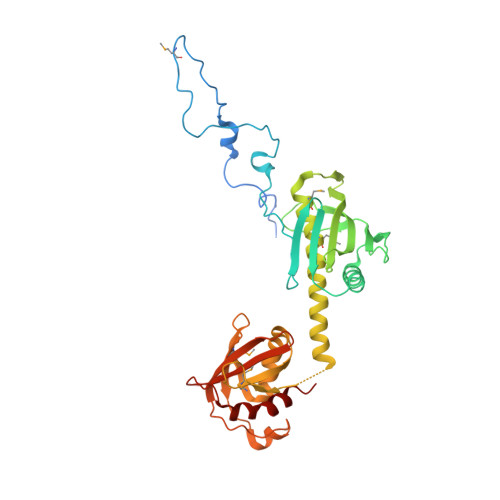
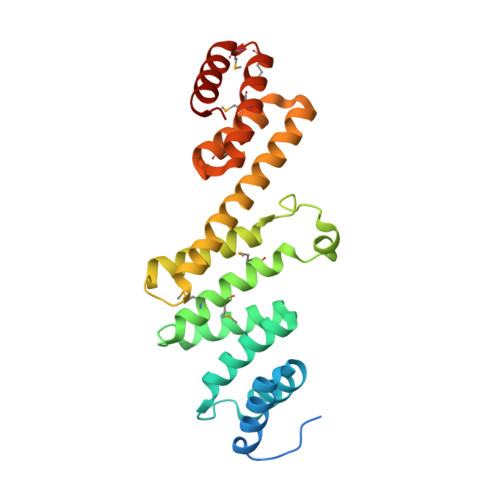
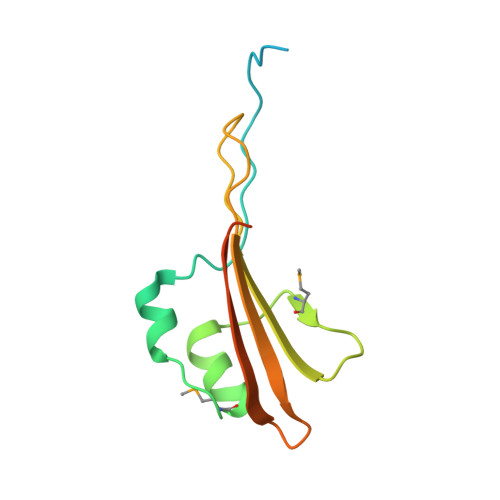
Structural basis of outer membrane protein insertion by the BAM complex.
Gu, Y., Li, H., Dong, H., Zeng, Y., Zhang, Z., Paterson, N.G., Stansfeld, P.J., Wang, Z., Zhang, Y., Wang, W., Dong, C.(2016) Nature 531: 47-52
- PubMed: 26901871
- DOI: https://doi.org/10.1038/nature17199
- Primary Citation of Related Structures:
5D0O, 5D0Q - PubMed Abstract:
All Gram-negative bacteria, mitochondria and chloroplasts have outer membrane proteins (OMPs) that perform many fundamental biological processes. The OMPs in Gram-negative bacteria are inserted and folded into the outer membrane by the β-barrel assembly machinery (BAM). The mechanism involved is poorly understood, owing to the absence of a structure of the entire BAM complex. Here we report two crystal structures of the Escherichia coli BAM complex in two distinct states: an inward-open state and a lateral-open state. Our structures reveal that the five polypeptide transport-associated domains of BamA form a ring architecture with four associated lipoproteins, BamB-BamE, in the periplasm. Our structural, functional studies and molecular dynamics simulations indicate that these subunits rotate with respect to the integral membrane β-barrel of BamA to induce movement of the β-strands of the barrel and promote insertion of the nascent OMP.
- Biomedical Research Centre, Norwich Medical School, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, UK.
Organizational Affiliation: