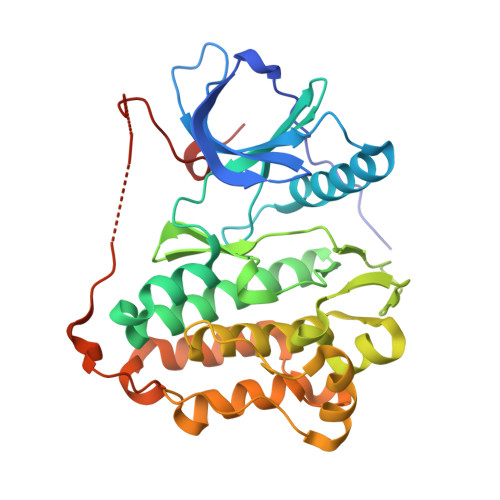
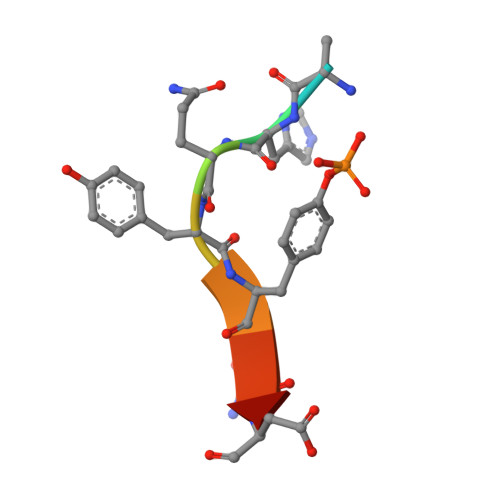
EGF-receptor specificity for phosphotyrosine-primed substrates provides signal integration with Src.
Begley, M.J., Yun, C.H., Gewinner, C.A., Asara, J.M., Johnson, J.L., Coyle, A.J., Eck, M.J., Apostolou, I., Cantley, L.C.(2015) Nat Struct Mol Biol 22: 983-990
- PubMed: 26551075
- DOI: https://doi.org/10.1038/nsmb.3117
- Primary Citation of Related Structures:
5CZH, 5CZI - PubMed Abstract:
Aberrant activation of the EGF receptor (EGFR) contributes to many human cancers by activating the Ras-MAPK pathway and other pathways. EGFR signaling is augmented by Src-family kinases, but the mechanism is poorly understood. Here, we show that human EGFR preferentially phosphorylates peptide substrates that are primed by a prior phosphorylation. Using peptides based on the sequence of the adaptor protein Shc1, we show that Src mediates the priming phosphorylation, thus promoting subsequent phosphorylation by EGFR. Importantly, the doubly phosphorylated Shc1 peptide binds more tightly than singly phosphorylated peptide to the Ras activator Grb2; this binding is a key step in activating the Ras-MAPK pathway. Finally, a crystal structure of EGFR in complex with a primed Shc1 peptide reveals the structural basis for EGFR substrate specificity. These results provide a molecular explanation for the integration of Src and EGFR signaling with downstream effectors such as Ras.
- Division of Signal Transduction, Beth Israel Deaconess Medical Center, Boston, Massachusetts, USA.
Organizational Affiliation: