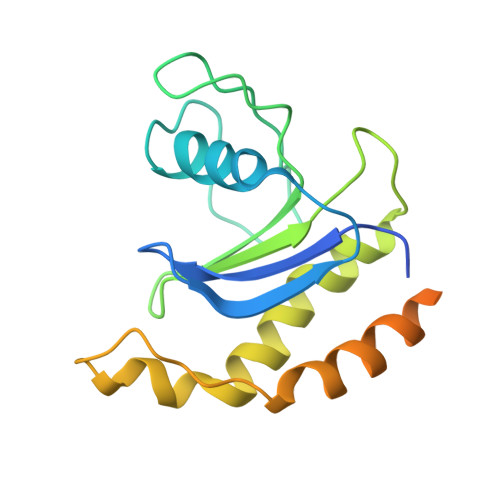
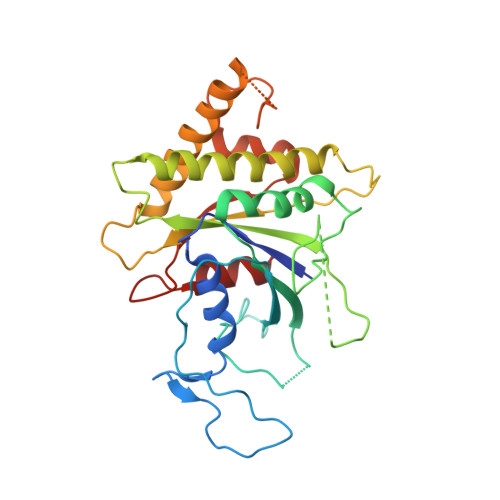
Structure and Switch Cycle of SR beta as Ancestral Eukaryotic GTPase Associated with Secretory Membranes.
Jadhav, B., Wild, K., Pool, M.R., Sinning, I.(2015) Structure 23: 1838-1847
- PubMed: 26299945
- DOI: https://doi.org/10.1016/j.str.2015.07.010
- Primary Citation of Related Structures:
5CK3, 5CK4, 5CK5 - PubMed Abstract:
G proteins of the Ras-family of small GTPases trace the evolution of eukaryotes. The earliest branching involves the closely related Arf, Sar1, and SRβ GTPases associated with secretory membranes. SRβ is an integral membrane component of the signal recognition particle (SRP) receptor that targets ribosome-nascent chain complexes to the ER. How SRβ integrates into the regulation of SRP-dependent membrane protein biogenesis is not known. Here we show that SRβ-GTP interacts with ribosomes only in presence of SRα and present crystal structures of SRβ in complex with the SRX domain of SRα in the GTP-bound state at 3.2 Å, and of GDP- and GDP · Mg(2+)-bound SRβ at 1.9 Å and 2.4 Å, respectively. We define the GTPase switch cycle of SRβ and identify specific differences to the Arf and Sar1 families with implications for GTPase regulation. Our data allow a better integration of SRβ into the scheme of protein targeting.
- Heidelberg University Biochemistry Center (BZH), Im Neuenheimer Feld 328, 69120 Heidelberg, Germany; Faculty of Life Sciences, University of Manchester, Oxford Road, Manchester M13 9PT, UK.
Organizational Affiliation: