The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division
Jia, M., Shan, Z., Yang, Y., Liu, C., Li, J., Luo, Z.G., Zhang, M., Cai, Y., Wen, W., Wang, W.(2015) Nat Commun 6: 8381-8381
- PubMed: 26423004
- DOI: https://doi.org/10.1038/ncomms9381
- Primary Citation of Related Structures:
5CFF - PubMed Abstract:
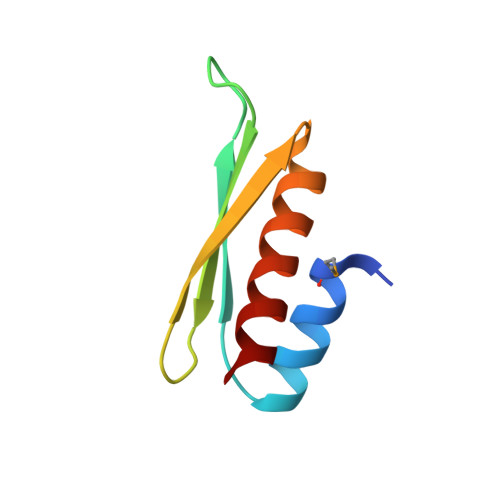
During the asymmetric division of Drosophila neuroblasts (NBs), the scaffold Miranda (Mira) coordinates the subcellular distribution of cell-fate determinants including Staufen (Stau) and segregates them into the ganglion mother cells (GMCs). Here we show the fifth double-stranded RNA (dsRNA)-binding domain (dsRBD5) of Stau is necessary and sufficient for binding to a coiled-coil region of Mira cargo-binding domain (CBD). The crystal structure of Mira514-595/Stau dsRBD5 complex illustrates that Mira forms an elongated parallel coiled-coil dimer, and two dsRBD5 symmetrically bind to the Mira dimer through their exposed β-sheet faces, revealing a previously unrecognized protein interaction mode for dsRBDs. We further demonstrate that the Mira-Stau dsRBD5 interaction is responsible for the asymmetric localization of Stau during Drosophila NB asymmetric divisions. Finally, we find the CBD-mediated dimer assembly is likely a common requirement for Mira to recognize and translocate other cargos including brain tumour (Brat).
- Shanghai Key Laboratory of Molecular Catalysis and Innovative Materials, Department of Chemistry and Key Laboratory of Molecular Medicine, Ministry of Education, Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, Shanghai 200433, China.
Organizational Affiliation: