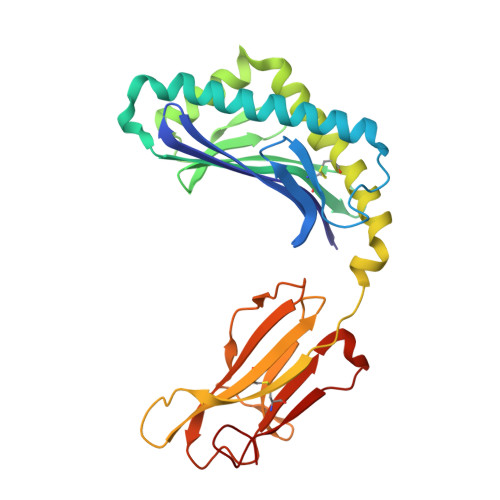
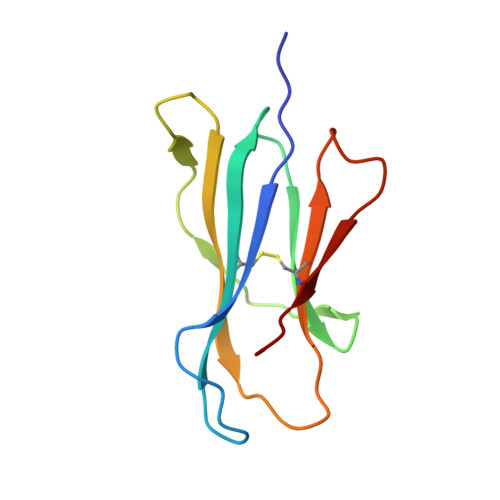
Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells.
Mansour, S., Tocheva, A.S., Cave-Ayland, C., Machelett, M.M., Sander, B., Lissin, N.M., Molloy, P.E., Baird, M.S., Stubs, G., Schroder, N.W., Schumann, R.R., Rademann, J., Postle, A.D., Jakobsen, B.K., Marshall, B.G., Gosain, R., Elkington, P.T., Elliott, T., Skylaris, C.K., Essex, J.W., Tews, I., Gadola, S.D.(2016) Proc Natl Acad Sci U S A 113: E1266-E1275
- PubMed: 26884207
- DOI: https://doi.org/10.1073/pnas.1519246113
- Primary Citation of Related Structures:
5C9J - PubMed Abstract:
Cluster of differentiation 1c (CD1c)-dependent self-reactive T cells are abundant in human blood, but self-antigens presented by CD1c to the T-cell receptors of these cells are poorly understood. Here we present a crystal structure of CD1c determined at 2.4 Å revealing an extended ligand binding potential of the antigen groove and a substantially different conformation compared with known CD1c structures. Computational simulations exploring different occupancy states of the groove reenacted these different CD1c conformations and suggested cholesteryl esters (CE) and acylated steryl glycosides (ASG) as new ligand classes for CD1c. Confirming this, we show that binding of CE and ASG to CD1c enables the binding of human CD1c self-reactive T-cell receptors. Hence, human CD1c adopts different conformations dependent on ligand occupancy of its groove, with CE and ASG stabilizing CD1c conformations that provide a footprint for binding of CD1c self-reactive T-cell receptors.
- Clinical and Experimental Sciences, Faculty of Medicine, University of Southampton, Southampton SO17 1BJ, United Kingdom; Institute for Life Sciences, University of Southampton, Southampton SO17 1BJ, United Kingdom; sgadola@gmail.com s.mansour@soton.ac.uk.
Organizational Affiliation: