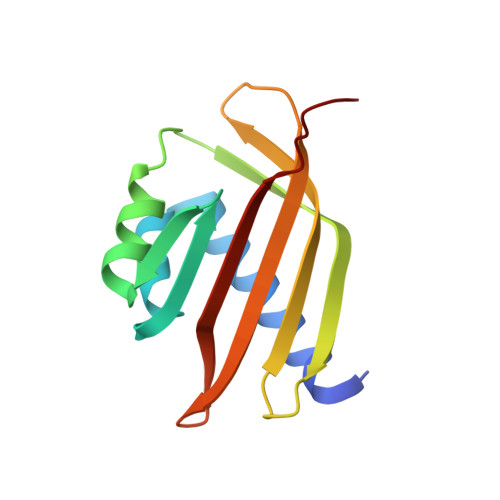
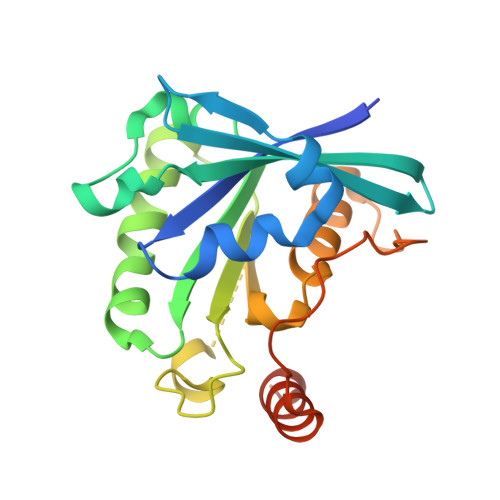
Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran.
Stewart, M., Kent, H.M., McCoy, A.J.(1998) J Mol Biology 277: 635-646
- PubMed: 9533885
- DOI: https://doi.org/10.1006/jmbi.1997.1602
- Primary Citation of Related Structures:
5BXQ - PubMed Abstract:
Nuclear transport factor 2 (NTF2) and the Ras-family GTPase Ran are two soluble components of the nuclear protein import machinery. NTF2 binds GDP-Ran selectively and this interaction is important for efficient nuclear protein import in vivo. We have used X-ray crystallography to determine the structure of the macromolecular complex formed between GDP-Ran and nuclear transport factor 2 (NTF2) at 2.5 A resolution. The interaction interface involves primarily the putative switch II loop of Ran (residues 65 to 78) and the hydrophobic cavity and surrounding surface of NTF2. The major contribution to the interaction made by the switch II loop accounts for the ability of NTF2 to discriminate between GDP and GTP-bound forms of Ran. The aromatic side-chain of Ran Phe72 inserts into the NTF2 cavity and accounts for 22% of the surface area buried by the interaction interface, while salt bridges are formed between Lys71 and Arg76 of Ran with Asp92/Asp94 and Glu42 of NTF2, respectively. These salt bridges account for the inhibition of the Ran-NTF2 interaction by NTF2 mutants such as E42 K and D92/94N in which the negatively charged residues surrounding the cavity were altered. Because the interaction interface maintains the positions of key Ran residues involved in binding MgGDP, NTF2 binding may help stabilize the switch state of Ran, possibly in the context of targeting it to other components of the nuclear protein import machinery to specify directionality of transport. The binding of GDP-Ran at the NTF2 cavity raises the possibility that this interaction might be modulated by a metabolite or small molecule substrate for NTF2's putative enzymatic activity.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge, CB2 2QH, England.
Organizational Affiliation: