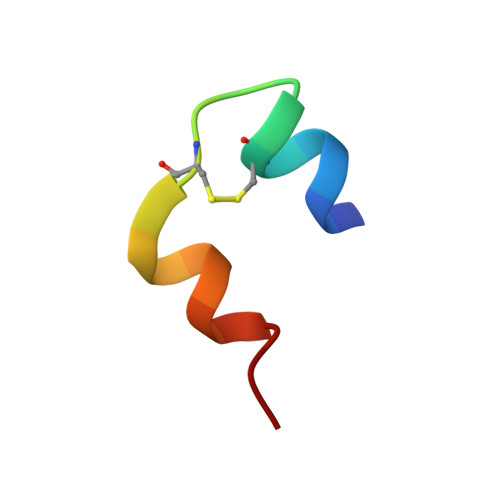
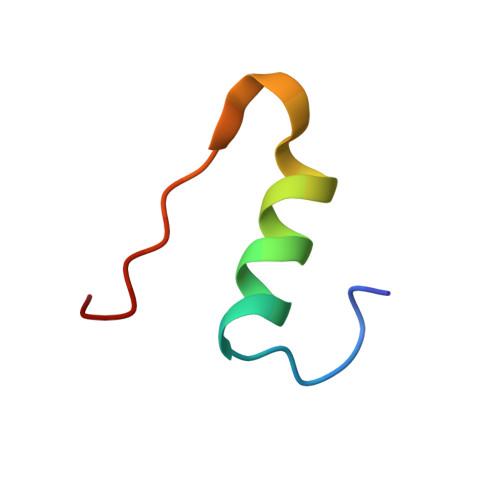
Structure, Aggregation, and Activity of a Covalent Insulin Dimer Formed During Storage of Neutral Formulation of Human Insulin.
Hjorth, C.F., Norrman, M., Wahlund, P.O., Benie, A.J., Petersen, B.O., Jessen, C.M., Pedersen, T.A., Vestergaard, K., Steensgaard, D.B., Pedersen, J.S., Naver, H., Hubalek, F., Poulsen, C., Otzen, D.(2016) J Pharm Sci 105: 1376-1386
- PubMed: 26921119
- DOI: https://doi.org/10.1016/j.xphs.2016.01.003
- Primary Citation of Related Structures:
5BTS - PubMed Abstract:
A specific covalently linked dimeric species of insulin high molecular weight products (HMWPs), formed during prolonged incubation of a neutral pharmaceutical formulation of human insulin, were characterized in terms of tertiary structure, self-association, biological activity, and fibrillation properties. The dimer was formed by a covalent link between A21Asn and B29Lys. It was analyzed using static and dynamic light scattering and small-angle X-ray scattering to evaluate its self-association behavior. The tertiary structure was obtained using nuclear magnetic resonance and X-ray crystallography. The biological activity of HMWP was determined using 2 in vitro assays, and its influence on fibrillation was investigated using Thioflavin T assays. The dimer's tertiary structure was nearly identical to that of the noncovalent insulin dimer, and it was able to form hexamers in the presence of zinc. The dimer exhibited reduced propensity for self-association in the absence of zinc but significantly postponed the onset of fibrillation in insulin formulations. Consistent with its dimeric state, the tested species of HMWP showed little to no biological activity in the used assays. This study is the first detailed characterization of a specific type of human insulin HMWP formed during storage of a marketed pharmaceutical formulation. These results indicate that this specific type of HMWP is unlikely to antagonize the physical stability of the formulation, as HMWP retained a tertiary structure similar to the noncovalent dimer and participated in hexamer assembly in the presence of zinc. In addition, increasing amounts of HMWP reduce the rate of insulin fibrillation.
- Diabetes Protein Engineering, Novo Nordisk A/S, 2760 Måløv, Denmark; Department of Molecular Biology and Genetics, Interdisciplinary Nanoscience Center (iNANO), Aarhus University, 8000 Aarhus C, Denmark. Electronic address: cfth@novonordisk.com.
Organizational Affiliation: