Template-dependent nucleotide addition in the reverse (3'-5') direction by Thg1-like protein
Kimura, S., Suzuki, T., Chen, M., Kato, K., Yu, J., Nakamura, A., Tanaka, I., Yao, M.(2016) Sci Adv 2: e1501397-e1501397
- PubMed: 27051866
- DOI: https://doi.org/10.1126/sciadv.1501397
- Primary Citation of Related Structures:
5AXK, 5AXL, 5AXM, 5AXN - PubMed Abstract:
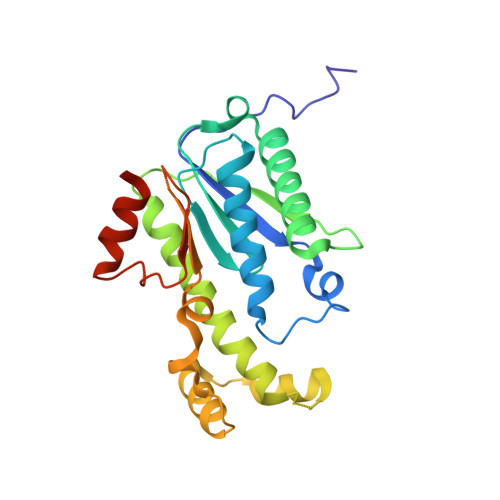
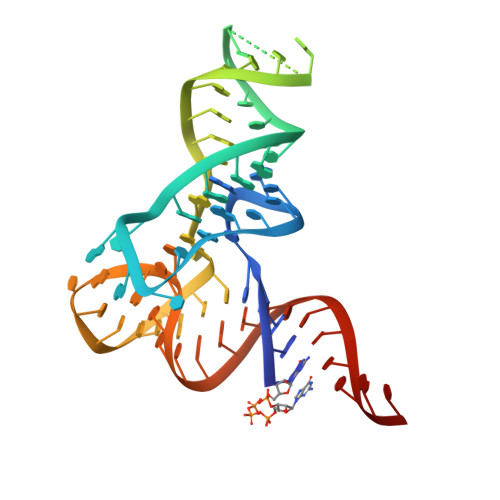
Thg1-like protein (TLP) catalyzes the addition of a nucleotide to the 5'-end of truncated transfer RNA (tRNA) species in a Watson-Crick template-dependent manner. The reaction proceeds in two steps: the activation of the 5'-end by adenosine 5'-triphosphate (ATP)/guanosine 5'-triphosphate (GTP), followed by nucleotide addition. Structural analyses of the TLP and its reaction intermediates have revealed the atomic detail of the template-dependent elongation reaction in the 3'-5' direction. The enzyme creates two substrate binding sites for the first- and second-step reactions in the vicinity of one reaction center consisting of two Mg(2+) ions, and the two reactions are executed at the same reaction center in a stepwise fashion. When the incoming nucleotide is bound to the second binding site with Watson-Crick hydrogen bonds, the 3'-OH of the incoming nucleotide and the 5'-triphosphate of the tRNA are moved to the reaction center where the first reaction has occurred. That the 3'-5' elongation enzyme performs this elaborate two-step reaction in one catalytic center suggests that these two reactions have been inseparable throughout the process of protein evolution. Although TLP and Thg1 have similar tetrameric organization, the tRNA binding mode of TLP is different from that of Thg1, a tRNA(His)-specific G-1 addition enzyme. Each tRNA(His) binds to three of the four Thg1 tetramer subunits, whereas in TLP, tRNA only binds to a dimer interface and the elongation reaction is terminated by measuring the accepter stem length through the flexible β-hairpin. Furthermore, mutational analyses show that tRNA(His) is bound to TLP in a similar manner as Thg1, thus indicating that TLP has a dual binding mode.
- Graduate School of Life Science, Hokkaido University, Sapporo 060-0810, Japan.
Organizational Affiliation: