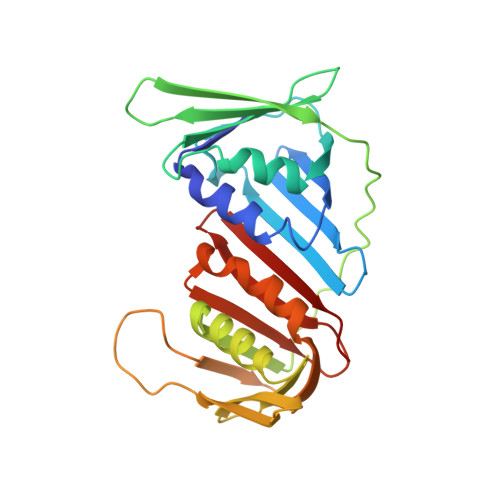
TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress.
Hoffmann, S., Smedegaard, S., Nakamura, K., Mortuza, G.B., Raschle, M., Ibanez de Opakua, A., Oka, Y., Feng, Y., Blanco, F.J., Mann, M., Montoya, G., Groth, A., Bekker-Jensen, S., Mailand, N.(2016) J Cell Biol 212: 63-75
- PubMed: 26711499
- DOI: https://doi.org/10.1083/jcb.201506071
- Primary Citation of Related Structures:
4ZTD - PubMed Abstract:
Cellular genomes are highly vulnerable to perturbations to chromosomal DNA replication. Proliferating cell nuclear antigen (PCNA), the processivity factor for DNA replication, plays a central role as a platform for recruitment of genome surveillance and DNA repair factors to replication forks, allowing cells to mitigate the threats to genome stability posed by replication stress. We identify the E3 ubiquitin ligase TRAIP as a new factor at active and stressed replication forks that directly interacts with PCNA via a conserved PCNA-interacting peptide (PIP) box motif. We show that TRAIP promotes ATR-dependent checkpoint signaling in human cells by facilitating the generation of RPA-bound single-stranded DNA regions upon replication stress in a manner that critically requires its E3 ligase activity and is potentiated by the PIP box. Consequently, loss of TRAIP function leads to enhanced chromosomal instability and decreased cell survival after replication stress. These findings establish TRAIP as a PCNA-binding ubiquitin ligase with an important role in protecting genome integrity after obstacles to DNA replication.
- Ubiquitin Signaling Group, The Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, 2200 Copenhagen, Denmark.
Organizational Affiliation: