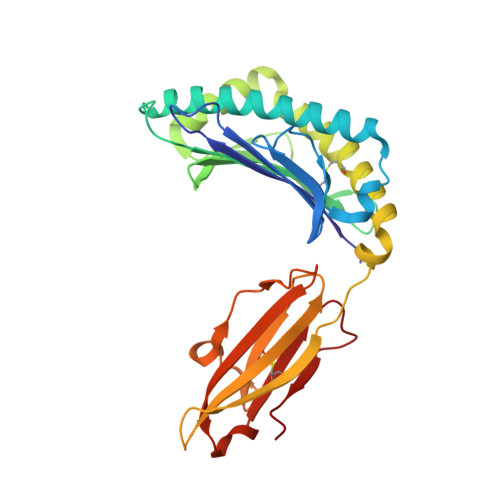
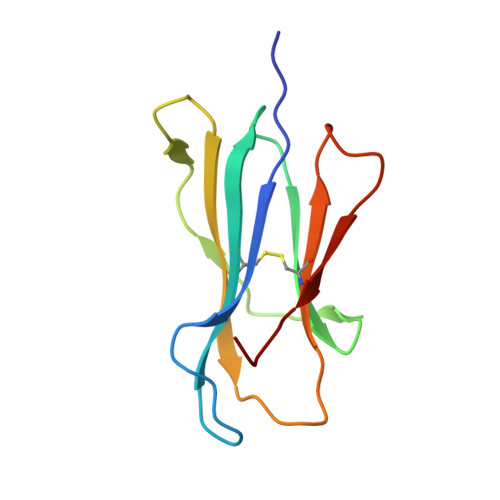
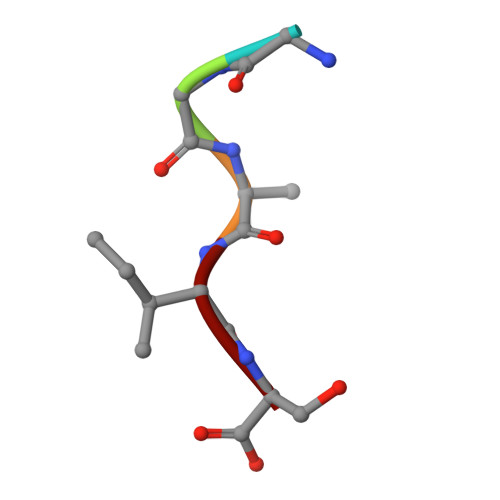
Crystal structure of the N-myristoylated lipopeptide-bound MHC class I complex
Morita, D., Yamamoto, Y., Mizutani, T., Ishikawa, T., Suzuki, J., Igarashi, T., Mori, N., Shiina, T., Inoko, H., Fujita, H., Iwai, K., Tanaka, Y., Mikami, B., Sugita, M.(2016) Nat Commun 7: 10356-10356
- PubMed: 26758274
- DOI: https://doi.org/10.1038/ncomms10356
- Primary Citation of Related Structures:
4ZFZ - PubMed Abstract:
The covalent conjugation of a 14-carbon saturated fatty acid (myristic acid) to the amino-terminal glycine residue is critical for some viral proteins to function. This protein lipidation modification, termed N-myristoylation, is targeted by host cytotoxic T lymphocytes (CTLs) that specifically recognize N-myristoylated short peptides; however, the molecular mechanisms underlying lipopeptide antigen (Ag) presentation remain elusive. Here we show that a primate major histocompatibility complex (MHC) class I-encoded protein is capable of binding N-myristoylated 5-mer peptides and presenting them to specific CTLs. A high-resolution X-ray crystallographic analysis of the MHC class I:lipopeptide complex reveals an Ag-binding groove that is elaborately constructed to bind N-myristoylated short peptides rather than prototypic 9-mer peptides. The identification of lipopeptide-specific, MHC class I-restricted CTLs indicates that the widely accepted concept of MHC class I-mediated presentation of long peptides to CTLs may need some modifications to incorporate a novel MHC class I function of lipopeptide Ag presentation.
- Laboratory of Cell Regulation, Institute for Virus Research, Kyoto University, 53 Kawahara-cho, Shogoin, Sakyo-ku, Kyoto 606-8507, Japan.
Organizational Affiliation: