Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
Chandran, T., Sharma, A., Vijayan, M.(2015) J Biosci 40: 929-941
- PubMed: 26648038
- DOI: https://doi.org/10.1007/s12038-015-9573-x
- Primary Citation of Related Structures:
4Z8S, 4Z9W, 4ZA3, 4ZBV, 4ZFU, 4ZFW, 4ZFY, 4ZGR, 4ZLB - PubMed Abstract:
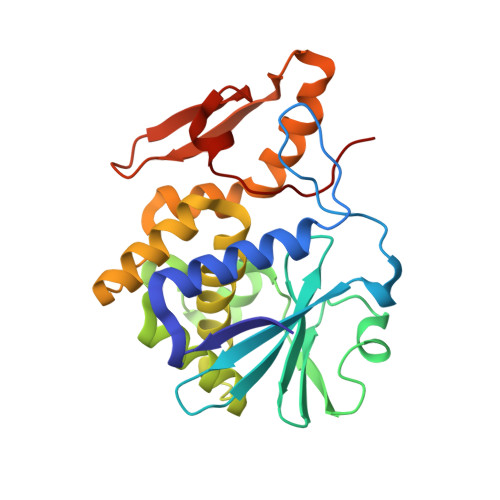
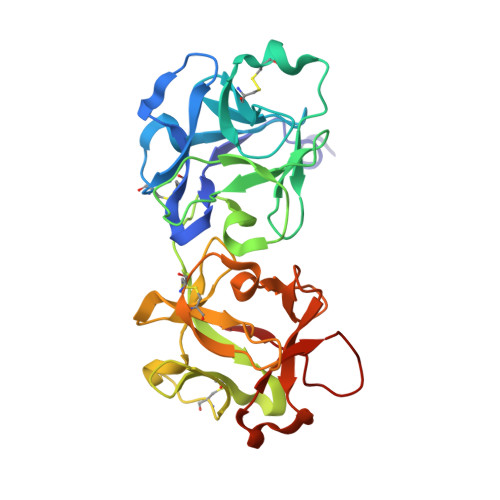
The structures of nine independent crystals of bitter gourd seed lectin (BGSL), a non-toxic homologue of type II RIPs, and its sugar complexes have been determined. The four-chain, two-fold symmetric, protein is made up of two identical two-chain modules, each consisting of a catalytic chain and a lectin chain, connected by a disulphide bridge. The lectin chain is made up of two domains. Each domain carries a carbohydrate binding site in type II RIPs of known structure. BGSL has a sugar binding site only on one domain, thus impairing its interaction at the cell surface. The adenine binding site in the catalytic chain is defective. Thus, defects in sugar binding as well as adenine binding appear to contribute to the non-toxicity of the lectin. The plasticity of the molecule is mainly caused by the presence of two possible well defined conformations of a surface loop in the lectin chain. One of them is chosen in the sugar complexes, in a case of conformational selection, as the chosen conformation facilitates an additional interaction with the sugar, involving an arginyl residue in the loop. The N-glycosylation of the lectin involves a plant-specific glycan while that in toxic type II RIPs of known structure involves a glycan which is animal as well as plant specific.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560012, India.
Organizational Affiliation: