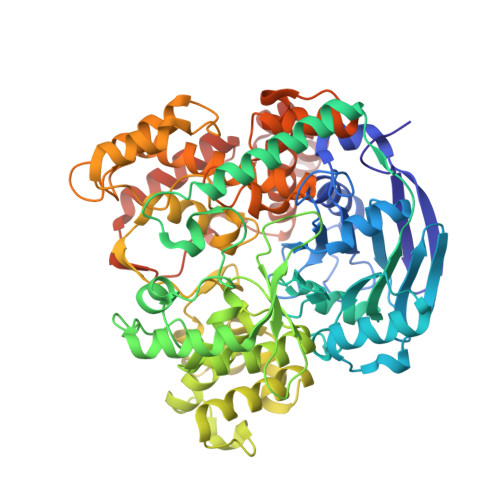
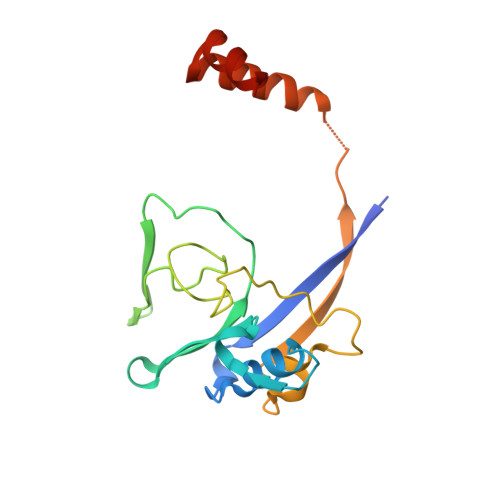
Structural basis of enzymatic benzene ring reduction.
Weinert, T., Huwiler, S.G., Kung, J.W., Weidenweber, S., Hellwig, P., Stark, H.J., Biskup, T., Weber, S., Cotelesage, J.J., George, G.N., Ermler, U., Boll, M.(2015) Nat Chem Biol 11: 586-591
- PubMed: 26120796
- DOI: https://doi.org/10.1038/nchembio.1849
- Primary Citation of Related Structures:
4Z3W, 4Z3X, 4Z3Y, 4Z3Z, 4Z40 - PubMed Abstract:
In chemical synthesis, the widely used Birch reduction of aromatic compounds to cyclic dienes requires alkali metals in ammonia as extremely low-potential electron donors. An analogous reaction is catalyzed by benzoyl-coenzyme A reductases (BCRs) that have a key role in the globally important bacterial degradation of aromatic compounds at anoxic sites. Because of the lack of structural information, the catalytic mechanism of enzymatic benzene ring reduction remained obscure. Here, we present the structural characterization of a dearomatizing BCR containing an unprecedented tungsten cofactor that transfers electrons to the benzene ring in an aprotic cavity. Substrate binding induces proton transfer from the bulk solvent to the active site by expelling a Zn(2+) that is crucial for active site encapsulation. Our results shed light on the structural basis of an electron transfer process at the negative redox potential limit in biology. They open the door for biological or biomimetic alternatives to a basic chemical synthetic tool.
- Max Planck Institute of Biophysics, Frankfurt, Germany.
Organizational Affiliation: