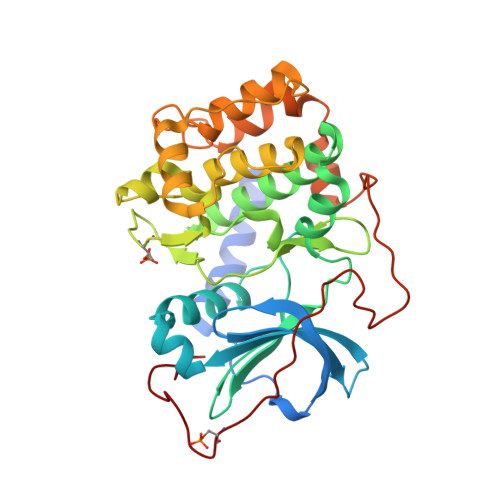
A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Pilger, J., Mazur, A., Monecke, P., Schreuder, H., Elshorst, B., Bartoschek, S., Langer, T., Schiffer, A., Krimm, I., Wegstroth, M., Lee, D., Hessler, G., Wendt, K.U., Becker, S., Griesinger, C.(2015) Angew Chem Int Ed Engl 54: 6511-6515
- PubMed: 25877959
- DOI: https://doi.org/10.1002/anie.201500671
- Primary Citation of Related Structures:
4C4X, 4C4Y, 4C4Z, 4YXR, 4YXS - PubMed Abstract:
Structure-based drug design (SBDD) is a powerful and widely used approach to optimize affinity of drug candidates. With the recently introduced INPHARMA method, the binding mode of small molecules to their protein target can be characterized even if no spectroscopic information about the protein is known. Here, we show that the combination of the spin-diffusion-based NMR methods INPHARMA, trNOE, and STD results in an accurate scoring function for docking modes and therefore determination of protein-ligand complex structures. Applications are shown on the model system protein kinase A and the drug targets glycogen phosphorylase and soluble epoxide hydrolase (sEH). Multiplexing of several ligands improves the reliability of the scoring function further. The new score allows in the case of sEH detecting two binding modes of the ligand in its binding site, which was corroborated by X-ray analysis.
- Abteilung für NMR-basierte Strukturbiologie, Max-Planck-Institut für biophysikalische Chemie, Am Fassberg 11, 37077 Göttingen (Germany).
Organizational Affiliation: