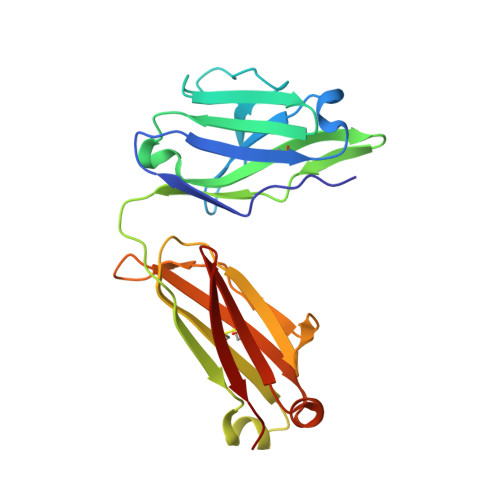
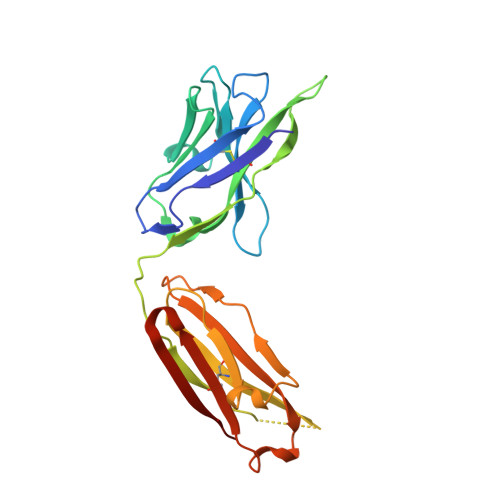
Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites.
Winarski, K.L., Thornburg, N.J., Yu, Y., Sapparapu, G., Crowe, J.E., Spiller, B.W.(2015) Proc Natl Acad Sci U S A 112: 9346-9351
- PubMed: 26170302
- DOI: https://doi.org/10.1073/pnas.1502762112
- Primary Citation of Related Structures:
4XNM, 4XNQ, 4XRC - PubMed Abstract:
Antigenic drift of circulating seasonal influenza viruses necessitates an international vaccine effort to reduce the impact on human health. A critical feature of the seasonal vaccine is that it stimulates an already primed immune system to diversify memory B cells to recognize closely related, but antigenically distinct, influenza glycoproteins (hemagglutinins). Influenza pandemics arise when hemagglutinins to which no preexisting adaptive immunity exists acquire the capacity to infect humans. Hemagglutinin 5 is one subtype to which little preexisting immunity exists and is only a few acquired mutations away from the ability to transmit efficiently between ferrets, and possibly humans. Here, we describe the structure and molecular mechanism of neutralization by H5.3, a vaccine-elicited antibody that neutralizes hemagglutinin 5 viruses and variants with expanded host range. H5.3 binds in the receptor-binding site, forming contacts that recapitulate many of the sialic acid interactions, as well as multiple peripheral interactions, yet is not sensitive to mutations that alter sialic acid binding. H5.3 is highly specific for a subset of H5 strains, and this specificity arises from interactions to the periphery of the receptor-binding site. H5.3 is also extremely potent, despite retaining germ line-like conformational flexibility.
- Department of Pathology, Microbiology and Immunology, Vanderbilt University School of Medicine, Nashville, TN 37232;
Organizational Affiliation: