Human Adenovirus 52 Uses Sialic Acid-containing Glycoproteins and the Coxsackie and Adenovirus Receptor for Binding to Target Cells.
Lenman, A., Liaci, A.M., Liu, Y., Ardahl, C., Rajan, A., Nilsson, E., Bradford, W., Kaeshammer, L., Jones, M.S., Frangsmyr, L., Feizi, T., Stehle, T., Arnberg, N.(2015) PLoS Pathog 11: e1004657-e1004657
- PubMed: 25674795
- DOI: https://doi.org/10.1371/journal.ppat.1004657
- Primary Citation of Related Structures:
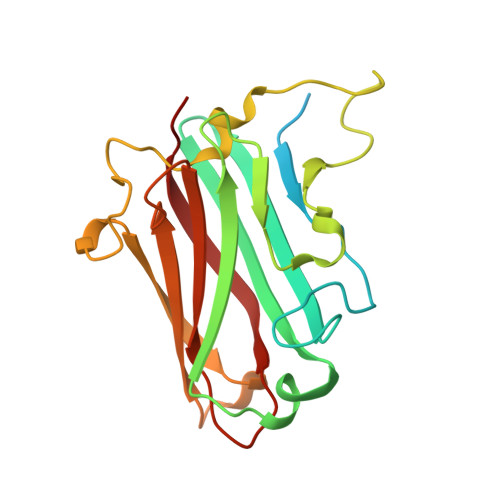
4XL8 - PubMed Abstract:
Most adenoviruses attach to host cells by means of the protruding fiber protein that binds to host cells via the coxsackievirus and adenovirus receptor (CAR) protein. Human adenovirus type 52 (HAdV-52) is one of only three gastroenteritis-causing HAdVs that are equipped with two different fiber proteins, one long and one short. Here we show, by means of virion-cell binding and infection experiments, that HAdV-52 can also attach to host cells via CAR, but most of the binding depends on sialylated glycoproteins. Glycan microarray, flow cytometry, surface plasmon resonance and ELISA analyses reveal that the terminal knob domain of the long fiber (52LFK) binds to CAR, and the knob domain of the short fiber (52SFK) binds to sialylated glycoproteins. X-ray crystallographic analysis of 52SFK in complex with 2-O-methylated sialic acid combined with functional studies of knob mutants revealed a new sialic acid binding site compared to other, known adenovirus:glycan interactions. Our findings shed light on adenovirus biology and may help to improve targeting of adenovirus-based vectors for gene therapy.
- Division of Virology, Department of Clinical Microbiology, Umeå University, Umeå, Sweden.
Organizational Affiliation: