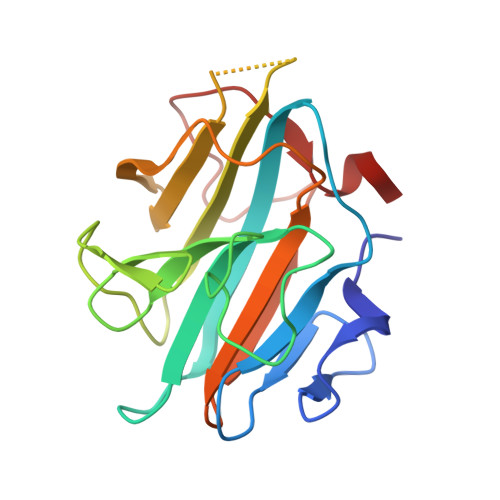
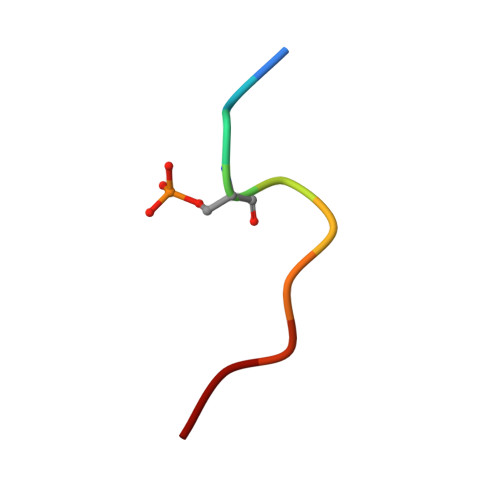
A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Zhang, P., Chaturvedi, C.P., Tremblay, V., Cramet, M., Brunzelle, J.S., Skiniotis, G., Brand, M., Shilatifard, A., Couture, J.F.(2015) Genes Dev 29: 123-128
- PubMed: 25593305
- DOI: https://doi.org/10.1101/gad.254870.114
- Primary Citation of Related Structures:
4X8N, 4X8P - PubMed Abstract:
The methyltransferase activity of the trithorax group (TrxG) protein MLL1 found within its COMPASS (complex associated with SET1)-like complex is allosterically regulated by a four-subunit complex composed of WDR5, RbBP5, Ash2L, and DPY30 (also referred to as WRAD). We report structural evidence showing that in WRAD, a concave surface of the Ash2L SPIa and ryanodine receptor (SPRY) domain binds to a cluster of acidic residues, referred to as the D/E box, in RbBP5. Mutational analysis shows that residues forming the Ash2L/RbBP5 interface are important for heterodimer formation, stimulation of MLL1 catalytic activity, and erythroid cell terminal differentiation. We also demonstrate that a phosphorylation switch on RbBP5 stimulates WRAD complex formation and significantly increases KMT2 (lysine [K] methyltransferase 2) enzyme methylation rates. Overall, our findings provide structural insights into the assembly of the WRAD complex and point to a novel regulatory mechanism controlling the activity of the KMT2/COMPASS family of lysine methyltransferases.
- Department of Biochemistry, Microbiology, and Immunology, Ottawa Institute of Systems Biology, University of Ottawa, Ottawa, Ontario K1H 8M5, Canada;
Organizational Affiliation: