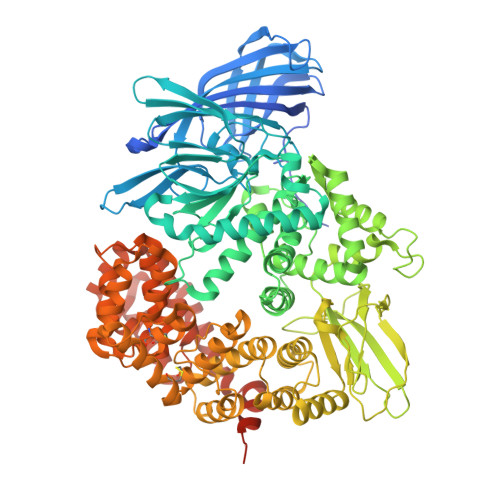
The Anopheles-midgut APN1 structure reveals a new malaria transmission-blocking vaccine epitope.
Atkinson, S.C., Armistead, J.S., Mathias, D.K., Sandeu, M.M., Tao, D., Borhani-Dizaji, N., Tarimo, B.B., Morlais, I., Dinglasan, R.R., Borg, N.A.(2015) Nat Struct Mol Biol 22: 532-539
- PubMed: 26075520
- DOI: https://doi.org/10.1038/nsmb.3048
- Primary Citation of Related Structures:
4WZ9 - PubMed Abstract:
Mosquito-based malaria transmission-blocking vaccines (mTBVs) target midgut-surface antigens of the Plasmodium parasite's obligate vector, the Anopheles mosquito. The alanyl aminopeptidase N (AnAPN1) is the leading mTBV immunogen; however, AnAPN1's role in Plasmodium infection of the mosquito and how anti-AnAPN1 antibodies functionally block parasite transmission have remained elusive. Here we present the 2.65-Å crystal structure of AnAPN1 and the immunoreactivity and transmission-blocking profiles of three monoclonal antibodies (mAbs) to AnAPN1, including mAb 4H5B7, which effectively blocks transmission of natural strains of Plasmodium falciparum. Using the AnAPN1 structure, we map the conformation-dependent 4H5B7 neoepitope to a previously uncharacterized region on domain 1 and further demonstrate that nonhuman-primate neoepitope-specific IgG also blocks parasite transmission. We discuss the prospect of a new biological function of AnAPN1 as a receptor for Plasmodium in the mosquito midgut and the implications for redesigning the AnAPN1 mTBV.
- Department of Biochemistry and Molecular Biology, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: