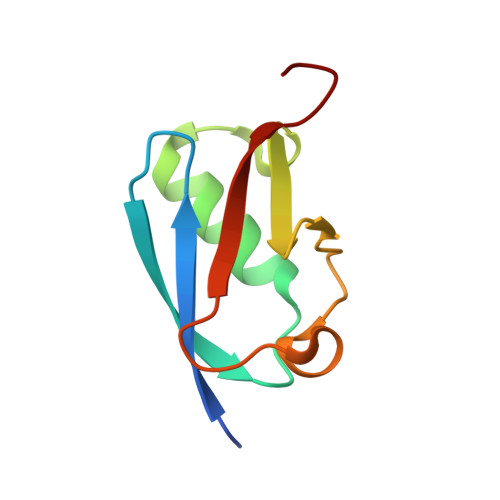
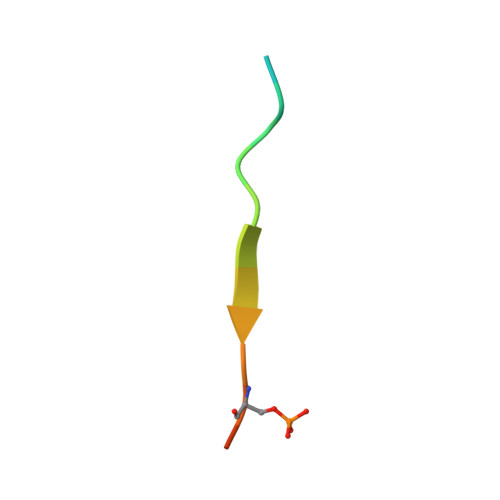
Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Cappadocia, L., Mascle, X.H., Bourdeau, V., Tremblay-Belzile, S., Chaker-Margot, M., Lussier-Price, M., Wada, J., Sakaguchi, K., Aubry, M., Ferbeyre, G., Omichinski, J.G.(2015) Structure 23: 126-138
- PubMed: 25497731
- DOI: https://doi.org/10.1016/j.str.2014.10.015
- Primary Citation of Related Structures:
4WJN, 4WJO, 4WJP, 4WJQ - PubMed Abstract:
PML and several other proteins localizing in PML-nuclear bodies (PML-NB) contain phosphoSIMs (SUMO-interacting motifs), and phosphorylation of this motif plays a key role in their interaction with SUMO family proteins. We examined the role that phosphorylation plays in the binding of the phosphoSIMs of PML and Daxx to SUMO1 at the atomic level. The crystal structures of SUMO1 bound to unphosphorylated and tetraphosphorylated PML-SIM peptides indicate that three phosphoserines directly contact specific positively charged residues of SUMO1. Surprisingly, the crystal structure of SUMO1 bound to a diphosphorylated Daxx-SIM peptide indicate that the hydrophobic residues of the phosphoSIM bind in a manner similar to that seen with PML, but important differences are observed when comparing the phosphorylated residues. Together, the results provide an atomic level description of how specific acetylation patterns within different SUMO family proteins can work together with phosphorylation of phosphoSIM's regions of target proteins to regulate binding specificity.
- Département de Biochimie et Médicine Moléculaire, Université de Montréal, C.P. 6128 Succursale Centre-Ville, Montréal, QC H3C 3J7, Canada.
Organizational Affiliation: