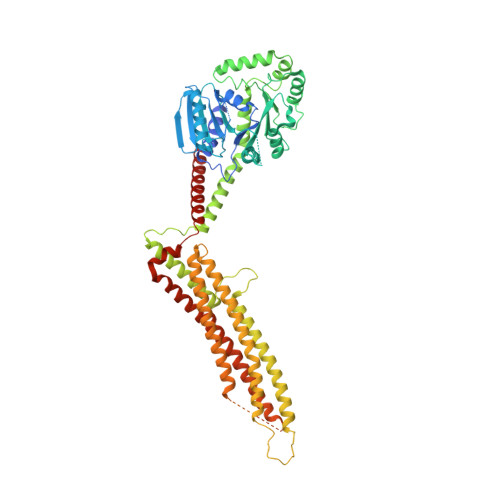
Structural Insight into HIV-1 Restriction by MxB.
Fribourgh, J.L., Nguyen, H.C., Matreyek, K.A., Alvarez, F.J., Summers, B.J., Dewdney, T.G., Aiken, C., Zhang, P., Engelman, A., Xiong, Y.(2014) Cell Host Microbe 16: 627-638
- PubMed: 25312384
- DOI: https://doi.org/10.1016/j.chom.2014.09.021
- Primary Citation of Related Structures:
4WHJ - PubMed Abstract:
The myxovirus resistance (Mx) proteins are interferon-induced dynamin GTPases that can inhibit a variety of viruses. Recently, MxB, but not MxA, was shown to restrict HIV-1 by an unknown mechanism that likely occurs in close proximity to the host cell nucleus and involves the viral capsid. Here, we present the crystal structure of MxB and reveal determinants involved in HIV-1 restriction. MxB adopts an extended antiparallel dimer and dimerization, but not higher-ordered oligomerization, is critical for restriction. Although MxB is structurally similar to MxA, the orientation of individual domains differs between MxA and MxB, and their antiviral functions rely on separate determinants, indicating distinct mechanisms for virus inhibition. Additionally, MxB directly binds the HIV-1 capsid, and this interaction depends on dimerization and the N terminus of MxB as well as the assembled capsid lattice. These insights establish a framework for understanding the mechanism by which MxB restricts HIV-1.
- Yale University, Molecular Biophysics and Biochemistry, New Haven, CT 06520, USA.
Organizational Affiliation: