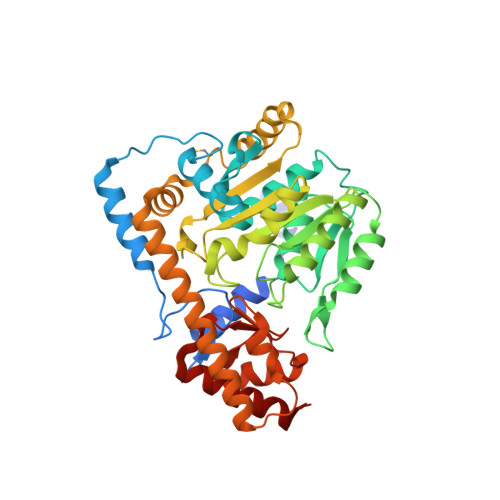
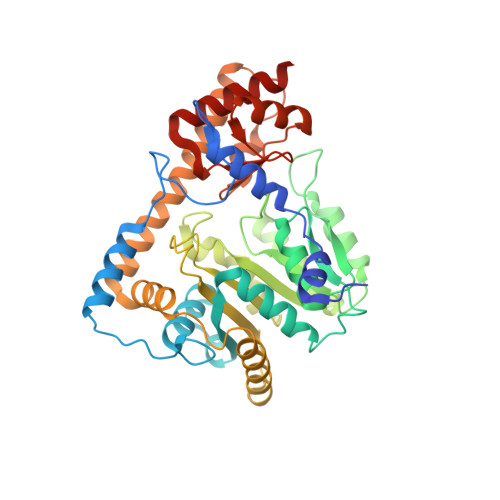
Structural basis for the relaxed substrate selectivity of Leishmania mexicana broad specificity aminotransferase.
Wen, J., Nowicki, C., Blankenfeldt, W.(2015) Mol Biochem Parasitol 202: 34-37
- PubMed: 26456583
- DOI: https://doi.org/10.1016/j.molbiopara.2015.09.007
- Primary Citation of Related Structures:
4WB0 - PubMed Abstract:
Leishmania species are early branching eukaryotic parasites that cause difficult-to-treat tissue-damaging diseases known as leishmaniases. As a hallmark of their parasitic lifestyle, Leishmaniae express a number of aminotransferases that are involved in important cellular processes and exhibit broader substrate specificity than their mammalian host's counterparts. Here, we have determined the crystal structure of the broad specificity aminotransferase from Leishmania mexicana (LmexBSAT) at 1.91Å resolution. LmexBSAT is a homodimer and belongs to the α-branch of family-I aminotransferases. Despite the fact that the protein was crystallized in the absence of substrates and has lost the pyridoxal-5'-phosphate (PLP) cofactor during crystallization, the structure resembles the closed, ligand-bound form of related enzymes such as chicken cytosolic aspartate aminotransferase. Its broader substrate specificity seems to be rooted in increased flexibility of a substrate-binding arginine (R291) and the interactions of this residue with the N-terminus of the second chain of the dimer.
- Technische Universität Dortmund, Fakultät Chemie, Otto-Hahn-Str. 6, 44227 Dortmund, Germany; Physical Biochemistry, Max Planck Institute for Molecular Physiology, Otto-Hahn-Str. 11, 44227 Dortmund, Germany.
Organizational Affiliation: