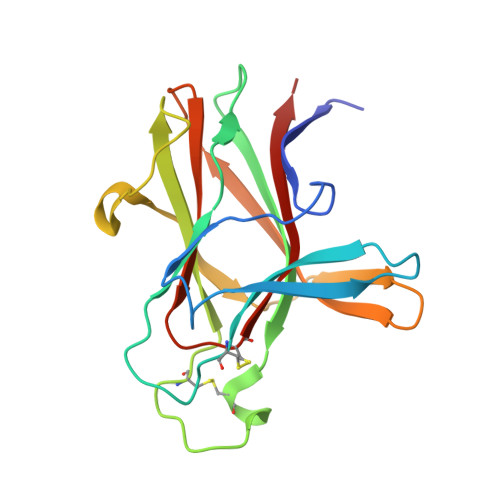
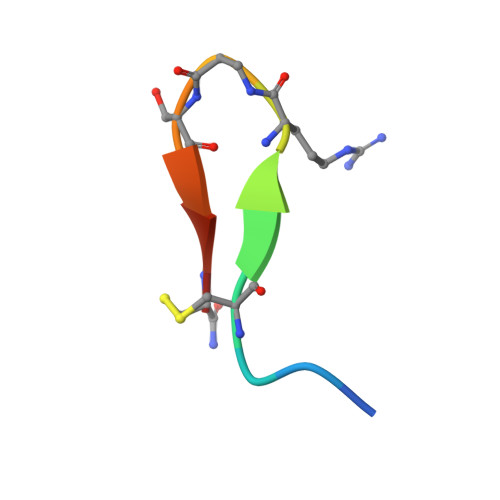
Development and Structural Analysis of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor.
Lamberto, I., Lechtenberg, B.C., Olson, E.J., Mace, P.D., Dawson, P.E., Riedl, S.J., Pasquale, E.B.(2014) ACS Chem Biol 9: 2787-2795
- PubMed: 25268696
- DOI: https://doi.org/10.1021/cb500677x
- Primary Citation of Related Structures:
4W4Z, 4W50 - PubMed Abstract:
The EphA4 receptor is highly expressed in the nervous system, and recent findings suggest that its signaling activity hinders neural repair and exacerbates certain neurodegenerative processes. EphA4 has also been implicated in cancer progression. Thus, EphA4 inhibitors represent potential therapeutic leads and useful research tools to elucidate the role of EphA4 in physiology and disease. Here, we report the structure of a cyclic peptide antagonist, APY, in complex with the EphA4 ligand-binding domain (LBD), which represents the first structure of a cyclic peptide bound to a receptor tyrosine kinase. The structure shows that the dodecameric APY efficiently occupies the ephrin ligand-binding pocket of EphA4 and promotes a "closed" conformation of the surrounding loops. Structure-guided relaxation of the strained APY β-turn and amidation of the C terminus to allow an additional intrapeptide hydrogen bond yielded APY-βAla8.am, an improved APY derivative that binds to EphA4 with nanomolar affinity. APY-βAla8.am potently inhibits ephrin-induced EphA4 activation in cells and EphA4-dependent neuronal growth cone collapse, while retaining high selectivity for EphA4. The two crystal structures of APY and APY-βAla8.am bound to EphA4, in conjunction with secondary phage display screens, highlighted peptide residues that are essential for EphA4 binding as well as residues that can be modified. Thus, the APY scaffold represents an exciting prototype, particularly since cyclic peptides have potentially favorable metabolic stability and are emerging as an important class of molecules for disruption of protein-protein interactions.
- Cancer Center, Sanford-Burnham Medical Research Institute , 10901 North Torrey Pines Road, La Jolla, California 92037, United States.
Organizational Affiliation: