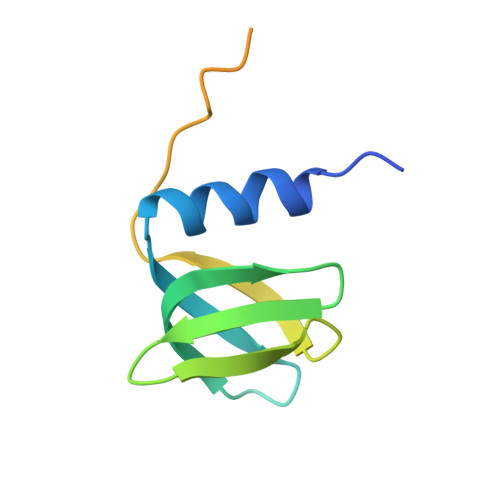
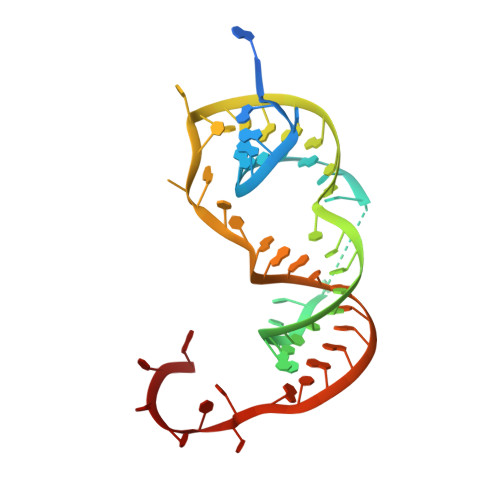
Recognition of the small regulatory RNA RydC by the bacterial Hfq protein.
Dimastrogiovanni, D., Frohlich, K.S., Bandyra, K.J., Bruce, H.A., Hohensee, S., Vogel, J., Luisi, B.F.(2014) Elife 3
- PubMed: 25551292
- DOI: https://doi.org/10.7554/eLife.05375
- Primary Citation of Related Structures:
4V2S - PubMed Abstract:
Bacterial small RNAs (sRNAs) are key elements of regulatory networks that modulate gene expression. The sRNA RydC of Salmonella sp. and Escherichia coli is an example of this class of riboregulators. Like many other sRNAs, RydC bears a 'seed' region that recognises specific transcripts through base-pairing, and its activities are facilitated by the RNA chaperone Hfq. The crystal structure of RydC in complex with E. coli Hfq at a 3.48 Å resolution illuminates how the protein interacts with and presents the sRNA for target recognition. Consolidating the protein-RNA complex is a host of distributed interactions mediated by the natively unstructured termini of Hfq. Based on the structure and other data, we propose a model for a dynamic effector complex comprising Hfq, small RNA, and the cognate mRNA target.
- Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom.
Organizational Affiliation: