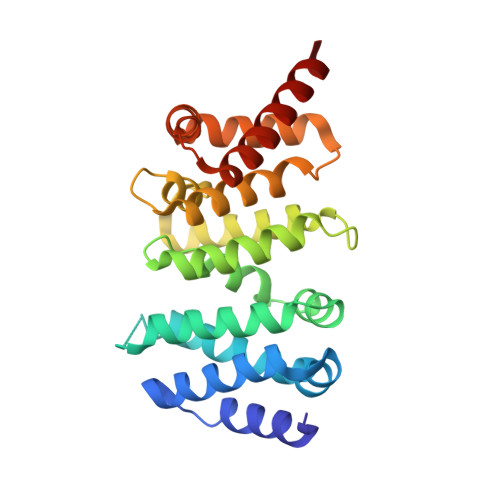
Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
Brenig, J., De Boor, S., Knyphausen, P., Kuhlmann, N., Wroblowski, S., Baldus, L., Scislowski, L., Artz, O., Trauschies, P., Baumann, U., Neundorf, I., Lammers, M.(2015) J Biological Chem 290: 14314
- PubMed: 25911102
- DOI: https://doi.org/10.1074/jbc.M114.621946
- Primary Citation of Related Structures:
4UWX - PubMed Abstract:
Diaphanous-related formins are eukaryotic actin nucleation factors regulated by an autoinhibitory interaction between the N-terminal RhoGTPase-binding domain (mDiaN) and the C-terminal Diaphanous-autoregulatory domain (DAD). Although the activation of formins by Rho proteins is well characterized, its inactivation is only marginally understood. Recently, liprin-α3 was shown to interact with mDia1. Overexpression of liprin-α3 resulted in a reduction of the cellular actin filament content. The molecular mechanisms of how liprin-α3 exerts this effect and counteracts mDia1 activation by RhoA are unknown. Here, we functionally and structurally define a minimal liprin-α3 core region, sufficient to recapitulate the liprin-α3 determined mDia1-respective cellular functions. We show that liprin-α3 alters the interaction kinetics and thermodynamics of mDiaN with RhoA·GTP and DAD. RhoA displaces liprin-α3 allosterically, whereas DAD competes with liprin-α3 for a highly overlapping binding site on mDiaN. Liprin-α3 regulates actin polymerization by lowering the regulatory potency of RhoA and DAD on mDiaN. We present a model of a mechanistically unexplored and new aspect of mDiaN regulation by liprin-α3.
- From the Institute for Genetics and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases (CECAD), Joseph-Stelzmann-Strasse 26, University of Cologne, 50931 Cologne, Germany and.
Organizational Affiliation: