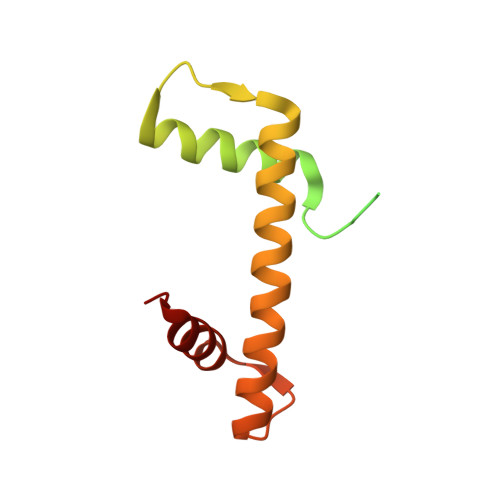
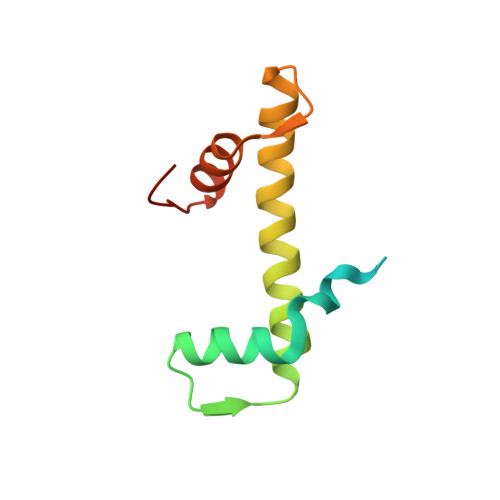
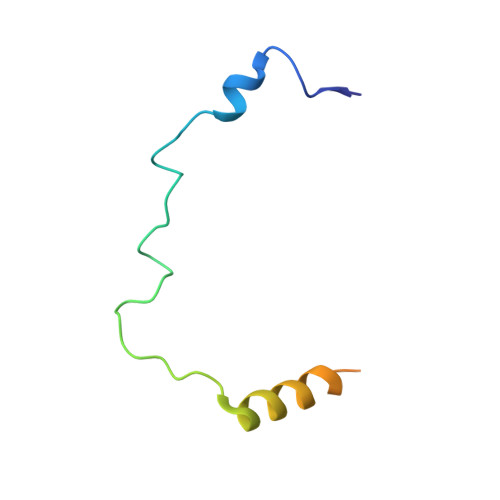
Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork.
Richet, N., Liu, D., Legrand, P., Velours, C., Corpet, A., Gaubert, A., Bakail, M., Moal-Raisin, G., Guerois, R., Compper, C., Besle, A., Guichard, B., Almouzni, G., Ochsenbein, F.(2015) Nucleic Acids Res 43: 1905
- PubMed: 25618846
- DOI: https://doi.org/10.1093/nar/gkv021
- Primary Citation of Related Structures:
4UUZ - PubMed Abstract:
MCM2 is a subunit of the replicative helicase machinery shown to interact with histones H3 and H4 during the replication process through its N-terminal domain. During replication, this interaction has been proposed to assist disassembly and assembly of nucleosomes on DNA. However, how this interaction participates in crosstalk with histone chaperones at the replication fork remains to be elucidated. Here, we solved the crystal structure of the ternary complex between the histone-binding domain of Mcm2 and the histones H3-H4 at 2.9 Å resolution. Histones H3 and H4 assemble as a tetramer in the crystal structure, but MCM2 interacts only with a single molecule of H3-H4. The latter interaction exploits binding surfaces that contact either DNA or H2B when H3-H4 dimers are incorporated in the nucleosome core particle. Upon binding of the ternary complex with the histone chaperone ASF1, the histone tetramer dissociates and both MCM2 and ASF1 interact simultaneously with the histones forming a 1:1:1:1 heteromeric complex. Thermodynamic analysis of the quaternary complex together with structural modeling support that ASF1 and MCM2 could form a chaperoning module for histones H3 and H4 protecting them from promiscuous interactions. This suggests an additional function for MCM2 outside its helicase function as a proper histone chaperone connected to the replication pathway.
- CEA, iBiTec-S, SB2SM, Laboratoire de Biologie Structurale et Radiobiologie, France Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Université Paris-Sud, Batiment 144, Gif-sur-Yvette, F-91191, France.
Organizational Affiliation: