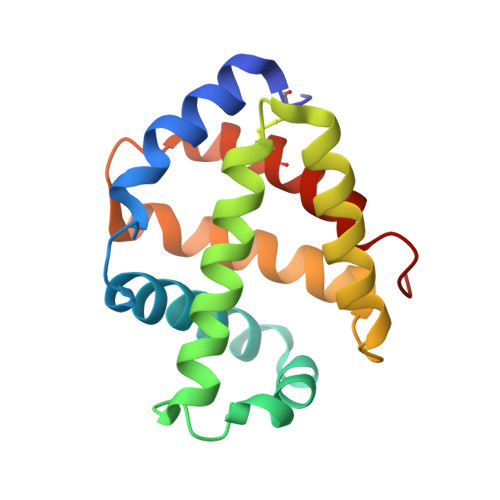
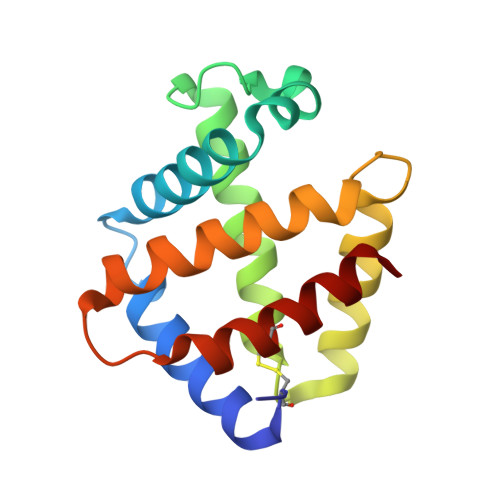
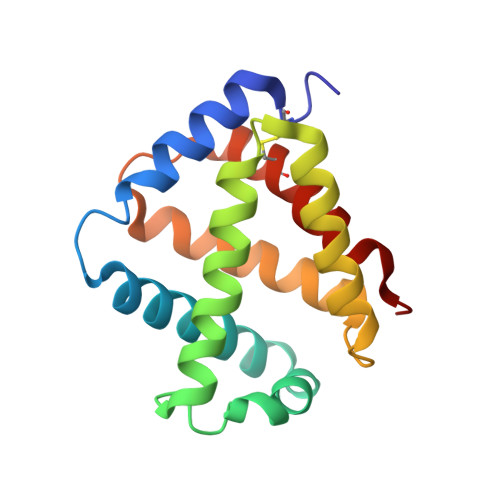
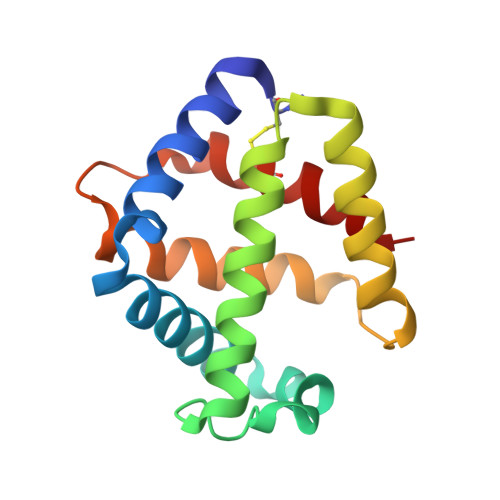
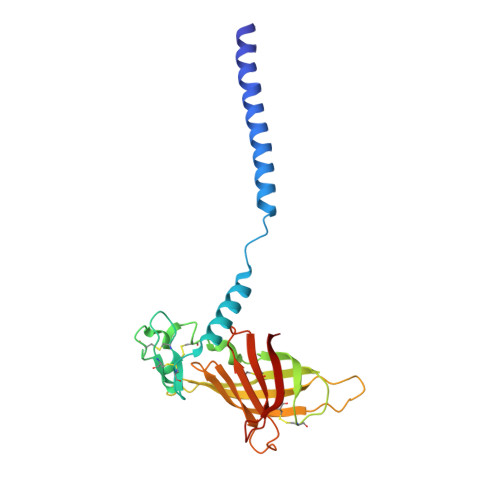
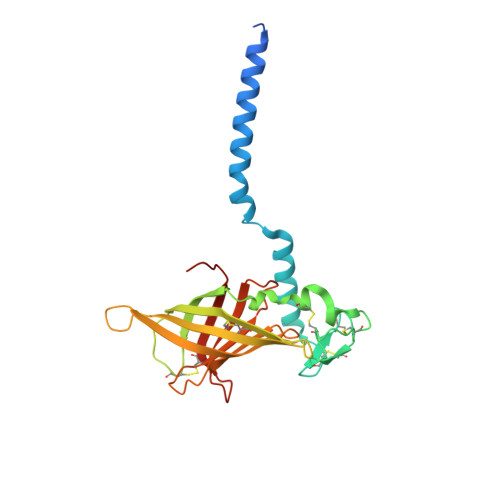
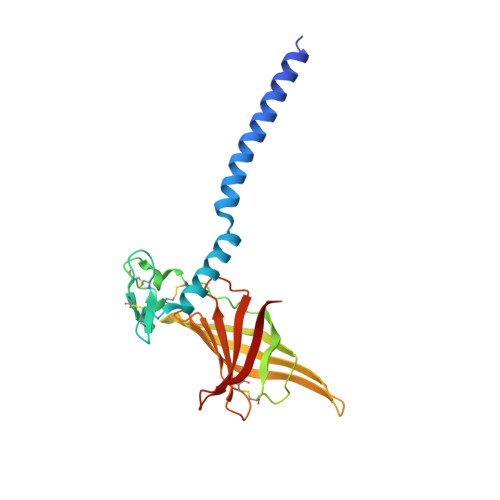
The structure of the giant haemoglobin from Glossoscolex paulistus.
Ruggiero Bachega, J.F., Vasconcelos Maluf, F., Andi, B., D'Muniz Pereira, H., Falsarella Carazzollea, M., Orville, A.M., Tabak, M., Brandao-Neto, J., Garratt, R.C., Horjales Reboredo, E.(2015) Acta Crystallogr D Biol Crystallogr 71: 1257-1271
- PubMed: 26057666
- DOI: https://doi.org/10.1107/S1399004715005453
- Primary Citation of Related Structures:
4U8U, 4WCH - PubMed Abstract:
The sequences of all seven polypeptide chains from the giant haemoglobin of the free-living earthworm Glossoscolex paulistus (HbGp) are reported together with the three-dimensional structure of the 3.6 MDa complex which they form. The refinement of the full particle, which has been solved at 3.2 Å resolution, the highest resolution reported to date for a hexagonal bilayer haemoglobin composed of 12 protomers, is reported. This has allowed a more detailed description of the contacts between subunits which are essential for particle stability. Interpretation of features in the electron-density maps suggests the presence of metal-binding sites (probably Zn(2+) and Ca(2+)) and glycosylation sites, some of which have not been reported previously. The former appear to be important for the integrity of the particle. The crystal structure of the isolated d chain (d-HbGp) at 2.1 Å resolution shows different interchain contacts between d monomers compared with those observed in the full particle. Instead of forming trimers, as seen in the complex, the isolated d chains associate to form dimers across a crystallographic twofold axis. These observations eliminate the possibility that trimers form spontaneously in solution as intermediates during the formation of the dodecameric globin cap and contribute to understanding of the possible ways in which the particle self-assembles.
- Instituto de Física de São Carlos, Universidade de São Paulo, São Carlos, Brazil.
Organizational Affiliation: