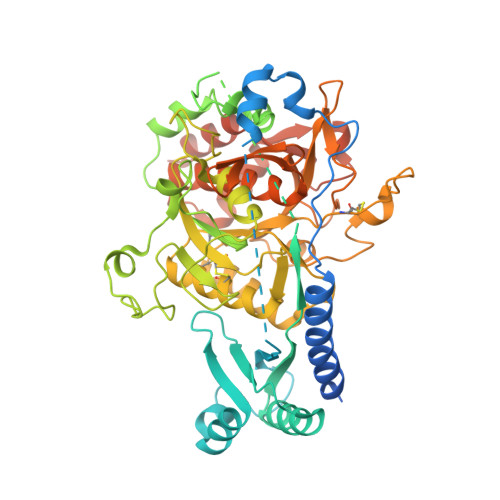
A novel Plasmodium-specific prodomain fold regulates the malaria drug target SUB1 subtilase.
Giganti, D., Bouillon, A., Tawk, L., Robert, F., Martinez, M., Crublet, E., Weber, P., Girard-Blanc, C., Petres, S., Haouz, A., Hernandez, J.F., Mercereau-Puijalon, O., Alzari, P.M., Barale, J.C.(2014) Nat Commun 5: 4833-4833
- PubMed: 25204226
- DOI: https://doi.org/10.1038/ncomms5833
- Primary Citation of Related Structures:
4TR2 - PubMed Abstract:
The Plasmodium subtilase SUB1 plays a pivotal role during the egress of malaria parasites from host hepatocytes and erythrocytes. Here we report the crystal structure of full-length SUB1 from the human-infecting parasite Plasmodium vivax, revealing a bacterial-like catalytic domain in complex with a Plasmodium-specific prodomain. The latter displays a novel architecture with an amino-terminal insertion that functions as a 'belt', embracing the catalytic domain to further stabilize the quaternary structure of the pre-protease, and undergoes calcium-dependent autoprocessing during subsequent activation. Although dispensable for recombinant enzymatic activity, the SUB1 'belt' could not be deleted in Plasmodium berghei, suggesting an essential role of this domain for parasite development in vivo. The SUB1 structure not only provides a valuable platform to develop new anti-malarial candidates against this promising drug target, but also defines the Plasmodium-specific 'belt' domain as a key calcium-dependent regulator of SUB1 during parasite egress from host cells.
- 1] Institut Pasteur, Unité de Microbiologie Structurale, Département de Biologie Structurale et Chimie, F-75015 Paris, France [2] CNRS UMR 3528, F-75015 Paris, France.
Organizational Affiliation: